Unravelling the Role of Rumen Microbial Communities, Genes, and Activities on Milk Fatty Acid Profile Using a Combination of Omics Approaches
- PMID: 33552010
- PMCID: PMC7859430
- DOI: 10.3389/fmicb.2020.590441
Unravelling the Role of Rumen Microbial Communities, Genes, and Activities on Milk Fatty Acid Profile Using a Combination of Omics Approaches
Abstract
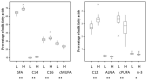
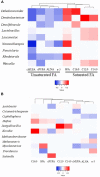
Milk products are an important component of human diets, with beneficial effects for human health, but also one of the major sources of nutritionally undesirable saturated fatty acids (SFA). Recent discoveries showing the importance of the rumen microbiome on dairy cattle health, metabolism and performance highlight that milk composition, and potentially milk SFA content, may also be associated with microorganisms, their genes and their activities. Understanding these mechanisms can be used for the development of cost-effective strategies for the production of milk with less SFA. This work aimed to compare the rumen microbiome between cows producing milk with contrasting FA profile and identify potentially responsible metabolic-related microbial mechanisms. Forty eight Holstein dairy cows were fed the same total mixed ration under the same housing conditions. Milk and rumen fluid samples were collected from all cows for the analysis of fatty acid profiles (by gas chromatography), the abundances of rumen microbiome communities and genes (by whole-genome-shotgun metagenomics), and rumen metabolome (using 500 MHz nuclear magnetic resonance). The following groups: (i) 24 High-SFA (66.9-74.4% total FA) vs. 24 Low-SFA (60.2-66.6%% total FA) cows, and (ii) 8 extreme High-SFA (69.9-74.4% total FA) vs. 8 extreme Low-SFA (60.2-64.0% total FA) were compared. Rumen of cows producing milk with more SFA were characterized by higher abundances of the lactic acid bacteria Lactobacillus, Leuconostoc, and Weissella, the acetogenic Proteobacteria Acetobacter and Kozakia, Mycobacterium, two fungi (Cutaneotrichosporon and Cyphellophora), and at a lesser extent Methanobrevibacter and the protist Nannochloropsis. Cows carrying genes correlated with milk FA also had higher concentrations of butyrate, propionate and tyrosine and lower concentrations of xanthine and hypoxanthine in the rumen. Abundances of rumen microbial genes were able to explain between 76 and 94% on the variation of the most abundant milk FA. Metagenomics and metabolomics analyses highlighted that cows producing milk with contrasting FA profile under the same diet, also differ in their rumen metabolic activities in relation to adaptation to reduced rumen pH, carbohydrate fermentation, and protein synthesis and metabolism.
Keywords: cow; metabolomics; metagenomics; microbial genes; milk fatty acids; rumen microbiome; rumen stress.
Copyright © 2021 Stergiadis, Cabeza-Luna, Mora-Ortiz, Stewart, Dewhurst, Humphries, Watson, Roehe and Auffret.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Agricultural and Food Research Council (1993). Energy and Protein Requirements of Ruminants. Wallingford, UK: CAB International.
-
- Aue W. P., Bartholdi E., Ernst R. R. (1975). Two-dimensional spectroscopy. application to nuclear magnetic resonance. J. Chem. Phys. 64:2229 10.1063/1.432450 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

