Quantifying the Stability of Coupled Genetic and Epigenetic Switches With Variational Methods
- PMID: 33552146
- PMCID: PMC7862759
- DOI: 10.3389/fgene.2020.636724
Quantifying the Stability of Coupled Genetic and Epigenetic Switches With Variational Methods
Abstract
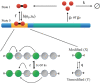
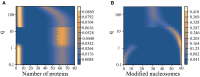
The Waddington landscape provides an intuitive metaphor to view development as a ball rolling down the hill, with distinct phenotypes as basins and differentiation pathways as valleys. Since, at a molecular level, cell differentiation arises from interactions among the genes, a mathematical definition for the Waddington landscape can, in principle, be obtained by studying the gene regulatory networks. For eukaryotes, gene regulation is inextricably and intimately linked to histone modifications. However, the impact of such modifications on both landscape topography and stability of attractor states is not fully understood. In this work, we introduced a minimal kinetic model for gene regulation that combines the impact of both histone modifications and transcription factors. We further developed an approximation scheme based on variational principles to solve the corresponding master equation in a second quantized framework. By analyzing the steady-state solutions at various parameter regimes, we found that histone modification kinetics can significantly alter the behavior of a genetic network, resulting in qualitative changes in gene expression profiles. The emerging epigenetic landscape captures the delicate interplay between transcription factors and histone modifications in driving cell-fate decisions.
Keywords: chromatin state; gene expression noise; gene network; minimum action; self-regulating gene.
Copyright © 2021 Sood and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Alexander F. J., Eyink G. L. (1997). Rayleigh-ritz calculation of effective potential far from equilibrium. Phys. Rev. Lett. 78, 1–4. 10.1103/PhysRevLett.78.1 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

