Genomic and transcriptomic resources for candidate gene discovery in the Ranunculids
- PMID: 33552749
- PMCID: PMC7845765
- DOI: 10.1002/aps3.11407
Genomic and transcriptomic resources for candidate gene discovery in the Ranunculids
Abstract
Premise: Multiple transitions from insect to wind pollination are associated with polyploidy and unisexual flowers in Thalictrum (Ranunculaceae), yet the underlying genetics remains unknown. We generated a draft genome of Thalictrum thalictroides, a representative of a clade with ancestral floral traits (diploid, hermaphrodite, and insect pollinated) and a model for functional studies. Floral transcriptomes of T. thalictroides and of wind-pollinated, andromonoecious T. hernandezii are presented as a resource to facilitate candidate gene discovery in flowers with different sexual and pollination systems.
Methods: A draft genome of T. thalictroides and two floral transcriptomes of T. thalictroides and T. hernandezii were obtained from HiSeq 2000 Illumina sequencing and de novo assembly.
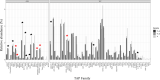
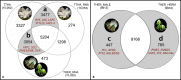
Results: The T. thalictroides de novo draft genome assembly consisted of 44,860 contigs (N50 = 12,761 bp, 243 Mbp total length) and contained 84.5% conserved embryophyte single-copy genes. Floral transcriptomes contained representatives of most eukaryotic core genes, and most of their genes formed orthogroups.
Discussion: To validate the utility of these resources, potential candidate genes were identified for the different floral morphologies using stepwise data set comparisons. Single-copy gene analysis and simple sequence repeat markers were also generated as a resource for population-level and phylogenetic studies.
Keywords: Ranunculaceae; Thalictrum hernandezii; Thalictrum thalictroides; draft genome; floral transcriptome; pollination syndrome; sexual system.
© 2021 Arias et al. Applications in Plant Sciences published by Wiley Periodicals LLC on behalf of Botanical Society of America.
Figures

 ) and hermaphrodite open flowers (
) and hermaphrodite open flowers ( ) occurring together in an inflorescence (B); inset, detail of young staminate flower. Scale bar = 1 cm.
) occurring together in an inflorescence (B); inset, detail of young staminate flower. Scale bar = 1 cm.
References
-
- Alexa, A. , and Rahnenfuhrer J.. 2010. topGO: Enrichment analysis for gene ontology. R Package Version 2: 2010.
-
- Andrews, S. 2015. FastQC: A quality control tool for high throughput sequence data, version 0.11.8. Website http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [accessed 18 December 2020].
-
- Arias, T. , Riaño‐Pachón D. M., and Di Stilio V. S.. 2020a. MISA results for transcriptomes (T. thalictroides and T. hernandezii) and genome (T. thalictroides) [published 12 June 2020]. Available at figshare 10.6084/m9.figshare.11984370.v8 [accessed 16 December 2020]. - DOI
-
- Arias, T. , and Riaño‐Pachón D. M.. 2020b. Interproscan results [published 14 May 2020]. Available at figshare 10.6084/m9.figshare.12302096.v1 [accessed 16 December 2020]. - DOI
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

