How enzymes harness highly unfavorable proton transfer reactions
- PMID: 33554401
- PMCID: PMC7980525
- DOI: 10.1002/pro.4037
How enzymes harness highly unfavorable proton transfer reactions
Abstract
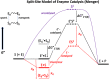
Acid-base reactions that are exceedingly unfavorable under standard conditions can be catalytically important at enzyme active sites. For example, in triose phosphate isomerase, a glutamate side chain (nominal pKa ≈ 4 in solution) can in fact deprotonate a CH group that is vicinal to a carbonyl (pKa ≈ 18 in solution). This is true because of three distinct interactions: (a) ground state pKa shifts due to environment polarity and electrostatics; (b) dramatic increases in effective molarity due to optimization of proximity and orientation; and (c) transition state pKa shifts due to binding interactions and the formation of strong low barrier hydrogen bonds. In this report, we review the literature showing that the sum of these three effects supplies more than enough free energy to push forward proton transfer reactions that under standard conditions are exceedingly nonspontaneous and slow.
Keywords: acid-base chemistry; activation energy; enzyme catalysis.
© 2021 The Protein Society.
Figures
References
-
- Menger FM. Enzyme reactivity from an organic perspective. Acc Chem Res. 1993;26:206–212.
-
- Radzicka A, Wolfenden R. A proficient enzyme. Science. 1995;267:90–93. - PubMed
-
- Menger FM. An alternative view of enzyme catalysis. Pure Appl Chem. 2005;77:1873–1886.
-
- Wolfenden R, Snider MJ. The depth of chemical time and the power of enzymes as catalysts. Acc Chem Res. 2001;34:938–945. - PubMed
-
- Kraut DA, Carroll KS, Herschlag D. Challenges in enzyme mechanism and energetics. Annu Rev Biochem. 2003;72:517–571. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

