Multimerization- and glycosylation-dependent receptor binding of SARS-CoV-2 spike proteins
- PMID: 33556147
- PMCID: PMC7895411
- DOI: 10.1371/journal.ppat.1009282
Multimerization- and glycosylation-dependent receptor binding of SARS-CoV-2 spike proteins
Abstract
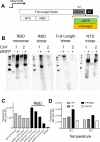
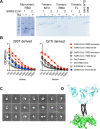
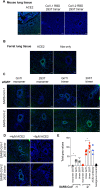
Receptor binding studies on sarbecoviruses would benefit from an available toolkit of recombinant spike proteins, or domains thereof, that recapitulate receptor binding properties of native viruses. We hypothesized that trimeric Receptor Binding Domain (RBD) proteins would be suitable candidates to study receptor binding properties of SARS-CoV-1 and -2. Here we created monomeric and trimeric fluorescent RBD proteins, derived from adherent HEK293T, as well as in GnTI-/- mutant cells, to analyze the effect of complex vs high mannose glycosylation on receptor binding. The results demonstrate that trimeric, complex glycosylated proteins are superior in receptor binding compared to monomeric and immaturely glycosylated variants. Although differences in binding to commonly used cell lines were minimal between the different RBD preparations, substantial differences were observed when respiratory tissues of experimental animals were stained. The RBD trimers demonstrated distinct ACE2 expression profiles in bronchiolar ducts and confirmed the higher binding affinity of SARS-CoV-2 over SARS-CoV-1. Our results show that complex glycosylated trimeric RBD proteins are attractive to analyze sarbecovirus receptor binding and explore ACE2 expression profiles in tissues.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

