Repair of DNA Double-Strand Breaks by the Nonhomologous End Joining Pathway
- PMID: 33556282
- PMCID: PMC8899865
- DOI: 10.1146/annurev-biochem-080320-110356
Repair of DNA Double-Strand Breaks by the Nonhomologous End Joining Pathway
Abstract
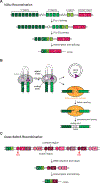
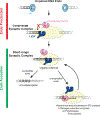
DNA double-strand breaks pose a serious threat to genome stability. In vertebrates, these breaks are predominantly repaired by nonhomologous end joining (NHEJ), which pairs DNA ends in a multiprotein synaptic complex to promote their direct ligation. NHEJ is a highly versatile pathway that uses an array of processing enzymes to modify damaged DNA ends and enable their ligation. The mechanisms of end synapsis and end processing have important implications for genome stability. Rapid and stable synapsis is necessary to limit chromosome translocations that result from the mispairing of DNA ends. Furthermore, end processing must be tightly regulated to minimize mutations at the break site. Here, we review our current mechanistic understanding of vertebrate NHEJ, with a particular focus on end synapsis and processing.
Keywords: DNA double-strand break; DNA end processing; DNA end synapsis; DNA repair; nonhomologous end joining.
Conflict of interest statement
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Figures

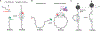


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

