Increased transcription and high translation efficiency lead to accumulation of androgen receptor splice variant after androgen deprivation therapy
- PMID: 33556543
- PMCID: PMC7940584
- DOI: 10.1016/j.canlet.2020.12.037
Increased transcription and high translation efficiency lead to accumulation of androgen receptor splice variant after androgen deprivation therapy
Abstract
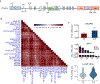
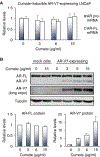
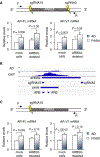
Upregulation of androgen receptor splice variants (AR-Vs), especially AR-V7, is associated with castration resistance of prostate cancer. At the RNA level, AR-V7 upregulation is generally coupled with increased full-length AR (AR-FL); consequently, AR-V7 and AR-Vs collectively constitute a minority of the AR population. However, Western blotting showed that the relative abundance of AR-V proteins is much higher in many castration-resistant prostate cancers (CRPCs). To address the mechanism underlying this discrepancy, we analyzed RNA-seq data from ~350 CRPC samples and found a positive correlation between all canonical and alternative AR splicing. This indicates that increased alternative splicing is not at the expense of canonical splicing. Instead, androgen deprivation releases AR-FL from repressing the transcription of the AR gene to induce coordinated increase of AR-FL and AR-V mRNAs. At the protein level, however, androgen deprivation induces AR-FL, but not AR-V, degradation. Moreover, AR-V7 is translated much faster than AR-FL. Thus, androgen-deprivation-induced AR-gene transcription and AR-FL protein decay, together with efficient AR-V7 translation, explain the discrepancy between the relative AR-V mRNA and protein abundances in many CRPCs, highlighting the inevitability of AR-V induction after endocrine therapy.
Keywords: AR negative autoregulation; AR-V mRNA stability; AR-V protein stability; AR-V translation; Castration resistance; Castration-resistant prostate cancer.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
Figures
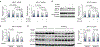

Similar articles
-
Rapid induction of androgen receptor splice variants by androgen deprivation in prostate cancer.Clin Cancer Res. 2014 Mar 15;20(6):1590-600. doi: 10.1158/1078-0432.CCR-13-1863. Epub 2014 Jan 21. Clin Cancer Res. 2014. PMID: 24449822 Free PMC article.
-
Growth Inhibition by Testosterone in an Androgen Receptor Splice Variant-Driven Prostate Cancer Model.Prostate. 2016 Dec;76(16):1536-1545. doi: 10.1002/pros.23238. Epub 2016 Jul 30. Prostate. 2016. PMID: 27473672
-
Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer.Cancer Res. 2012 Jul 15;72(14):3457-62. doi: 10.1158/0008-5472.CAN-11-3892. Epub 2012 Jun 18. Cancer Res. 2012. PMID: 22710436 Free PMC article.
-
Prognostic Value of Androgen Receptor Splice Variant 7 in the Treatment of Castration-resistant Prostate Cancer with Next generation Androgen Receptor Signal Inhibition: A Systematic Review and Meta-analysis.Eur Urol Focus. 2018 Jul;4(4):529-539. doi: 10.1016/j.euf.2017.01.004. Epub 2017 Jan 23. Eur Urol Focus. 2018. PMID: 28753843
-
Role of androgen receptor splice variants, their clinical relevance and treatment options.World J Urol. 2020 Mar;38(3):647-656. doi: 10.1007/s00345-018-02619-0. Epub 2019 Jan 19. World J Urol. 2020. PMID: 30659302 Review.
Cited by
-
Androgen receptor splice variant 7 functions independently of the full length receptor in prostate cancer cells.Cancer Lett. 2021 Oct 28;519:172-184. doi: 10.1016/j.canlet.2021.07.013. Epub 2021 Jul 10. Cancer Lett. 2021. PMID: 34256096 Free PMC article.
-
A narrative review of the role of glucocorticoid receptors in prostate cancer: developments in last 5 years.Transl Androl Urol. 2022 Aug;11(8):1189-1199. doi: 10.21037/tau-22-501. Transl Androl Urol. 2022. PMID: 36092840 Free PMC article. Review.
-
Synergistic Strategies for Castration-Resistant Prostate Cancer: Targeting AR-V7, Exploring Natural Compounds, and Optimizing FDA-Approved Therapies.Cancers (Basel). 2024 Aug 6;16(16):2777. doi: 10.3390/cancers16162777. Cancers (Basel). 2024. PMID: 39199550 Free PMC article. Review.
-
AR and PI3K/AKT in Prostate Cancer: A Tale of Two Interconnected Pathways.Int J Mol Sci. 2023 Jan 20;24(3):2046. doi: 10.3390/ijms24032046. Int J Mol Sci. 2023. PMID: 36768370 Free PMC article. Review.
-
Inhibiting androgen receptor splice variants with cysteine-selective irreversible covalent inhibitors to treat prostate cancer.Proc Natl Acad Sci U S A. 2023 Jan 3;120(1):e2211832120. doi: 10.1073/pnas.2211832120. Epub 2022 Dec 28. Proc Natl Acad Sci U S A. 2023. PMID: 36577061 Free PMC article.
References
-
- Egan A, Dong Y, Zhang H, Qi Y, Balk SP, Sartor O, Castration-resistant prostate cancer: adaptive responses in the androgen axis, Cancer Treat Rev, 40 (2014) 426–433. - PubMed
-
- Huang H, Tindall DJ, The role of the androgen receptor in prostate cancer, Crit Rev Eukaryot Gene Expr, 12 (2002) 193–207. - PubMed
-
- Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, Chen H, Kong X, Melamed J, Tepper CG, Kung HJ, Brodie AM, Edwards J, Qiu Y, A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth, Cancer Res, 69 (2009) 2305–2313. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

