Comparison of Antiviral Activity of Gemcitabine with 2'-Fluoro-2'-Deoxycytidine and Combination Therapy with Remdesivir against SARS-CoV-2
- PMID: 33557278
- PMCID: PMC7915419
- DOI: 10.3390/ijms22041581
Comparison of Antiviral Activity of Gemcitabine with 2'-Fluoro-2'-Deoxycytidine and Combination Therapy with Remdesivir against SARS-CoV-2
Abstract
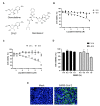
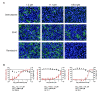
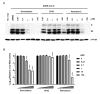
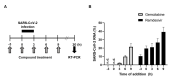
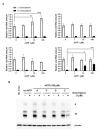
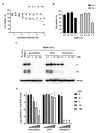
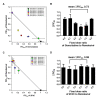
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the coronavirus disease 2019 (COVID-19) pandemic. The virus still spreads globally through human-to-human transmission. Nevertheless, there are no specific treatments clinically approved. This study aimed to compare antiviral activity of gemcitabine and its analogue 2'-fluoro-2'-deoxycytidine (2FdC) against SARS-CoV-2 as well as cytotoxicity in vitro. Fluorescent image-based antiviral assays revealed that gemcitabine was highly potent, with a 50% effective concentration (EC50) of 1.2 μM, more active than the well-known nucleoside monophosphate remdesivir (EC50 = 35.4 μM). In contrast, 2FdC was marginally active (EC50 = 175.2 μM). For all three compounds, the 50% cytotoxic concentration (CC50) values were over 300 μM toward Vero CCL-81 cells. Western blot and quantitative reverse-transcription polymerase chain reaction analyses verified that gemcitabine blocked viral protein expression in virus-infected cells, not only Vero CCL-81 cells but also Calu-3 human lung epithelial cells in a dose-dependent manner. It was found that gemcitabine has a synergistic effect when combined with remdesivir. This report suggests that the difluoro group of gemcitabine is critical for the antiviral activity and that its combination with other evaluated antiviral drugs, such as remdesivir, could be a desirable option to treat SARS-CoV-2 infection.
Keywords: 2′-fluoro-2′-deoxycytidine; SARS-CoV-2; antiviral activity; gemcitabine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Maters P.S., Perlman S. Coronaviridae. In: Knife D.M., Howley P.M., editors. Fields Virology. Volume 2. Lippincott Williams and Wilkins; Philadelphia, PA, USA: 2013. pp. 825–858.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

