Disruption of alpha-tubulin releases carbon catabolite repression and enhances enzyme production in Trichoderma reesei even in the presence of glucose
- PMID: 33557925
- PMCID: PMC7869464
- DOI: 10.1186/s13068-021-01887-0
Disruption of alpha-tubulin releases carbon catabolite repression and enhances enzyme production in Trichoderma reesei even in the presence of glucose
Abstract
Background: Trichoderma reesei is a filamentous fungus that is important as an industrial producer of cellulases and hemicellulases due to its high secretion of these enzymes and outstanding performance in industrial fermenters. However, the reduction of enzyme production caused by carbon catabolite repression (CCR) has long been a problem. Disruption of a typical transcriptional regulator, Cre1, does not sufficiently suppress this reduction in the presence of glucose.
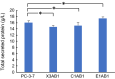
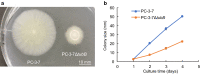
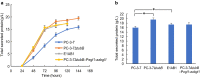
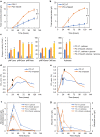
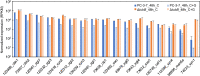
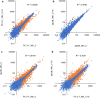
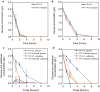
Results: We found that deletion of an α-tubulin (tubB) in T. reesei enhanced both the amount and rate of secretory protein production. Also, the tubulin-disrupted (ΔtubB) strain had high enzyme production and the same enzyme profile even if the strain was cultured in a glucose-containing medium. From transcriptome analysis, the ΔtubB strain exhibited upregulation of both cellulase and hemicellulase genes including some that were not originally induced by cellulose. Moreover, cellobiose transporter genes and the other sugar transporter genes were highly upregulated, and simultaneous uptake of glucose and cellobiose was also observed in the ΔtubB strain. These results suggested that the ΔtubB strain was released from CCR.
Conclusion: Trichoderma reesei α-tubulin is involved in the transcription of cellulase and hemicellulase genes, as well as in CCR. This is the first report of overcoming CCR by disrupting α-tubulin gene in T. reesei. The disruption of α-tubulin is a promising approach for creating next-generation enzyme-producing strains of T. reesei.
Keywords: Alpha-tubulin; Biomass saccharification enzyme; Carbon catabolite repression; Cellulase; Glucose resistant; Hemicellulase; Trichoderma reesei.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Nagesh M. Industrial Enzymes Market by Type (Carbohydrases, Proteases, Lipases, Polymerases & Nucleases, Other Types), Source, Application (Food & Beverages, Feed, Bioethanol, Detergents, Pulp & Paper, Textiles & Leather, Wastewater Treatment, Other Applications), Form, and Region - Global Forecast to 2026. MarketsandMarkets; 2020.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

