Finding Order in the Chaos: Outstanding Questions in Klebsiella pneumoniae Pathogenesis
- PMID: 33558323
- PMCID: PMC8090965
- DOI: 10.1128/IAI.00693-20
Finding Order in the Chaos: Outstanding Questions in Klebsiella pneumoniae Pathogenesis
Abstract
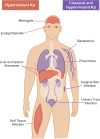
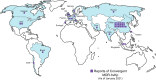
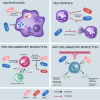
Klebsiella pneumoniae are Gram-negative facultative anaerobes that are found within host-associated commensal microbiomes, but they can also cause a wide range of infections that are often difficult to treat. These infections are caused by different pathotypes of K. pneumoniae, called either classical or hypervirulent strains. These two groups are genetically distinct, inhabit nonoverlapping geographies, and cause different types of harmful infections in humans. These distinct bacterial groups have also been found to interact differently with the host immune system. Initial innate immune defenses against K. pneumoniae infection include complement, macrophages, neutrophils, and monocytes; these defenses are primary strategies employed by the host to clear infections. K. pneumoniae pathogenesis depends upon the interactions between the microbe and each of these host defenses, and it is becoming increasingly apparent that bacterial genetic diversity impacts the outcomes of these interactions. Here, we highlight recent advances in our understanding of K. pneumoniae pathogenesis, with a focus on how bacterial evolution and diversity impact K. pneumoniae interactions with mammalian innate immune host defenses. We also discuss outstanding questions regarding how K. pneumoniae can frustrate normal immune responses, capitalize upon states of immunocompromise, and cause infections with high mortality.
Keywords: Klebsiella; evolution; innate immune system; pathogenesis.
Copyright © 2021 Gonzalez-Ferrer et al.
Figures

References
-
- Brisse S, Grimont PAD. 2006. The genus Klebsiella, p 159–196. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The prokaryotes, vol 3. Springer Nature, New York, NY.
-
- Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 112:E3574–E3581. 10.1073/pnas.1501049112. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

