Viral vector platforms within the gene therapy landscape
- PMID: 33558455
- PMCID: PMC7868676
- DOI: 10.1038/s41392-021-00487-6
Viral vector platforms within the gene therapy landscape
Abstract
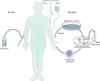
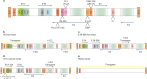
Throughout its 40-year history, the field of gene therapy has been marked by many transitions. It has seen great strides in combating human disease, has given hope to patients and families with limited treatment options, but has also been subject to many setbacks. Treatment of patients with this class of investigational drugs has resulted in severe adverse effects and, even in rare cases, death. At the heart of this dichotomous field are the viral-based vectors, the delivery vehicles that have allowed researchers and clinicians to develop powerful drug platforms, and have radically changed the face of medicine. Within the past 5 years, the gene therapy field has seen a wave of drugs based on viral vectors that have gained regulatory approval that come in a variety of designs and purposes. These modalities range from vector-based cancer therapies, to treating monogenic diseases with life-altering outcomes. At present, the three key vector strategies are based on adenoviruses, adeno-associated viruses, and lentiviruses. They have led the way in preclinical and clinical successes in the past two decades. However, despite these successes, many challenges still limit these approaches from attaining their full potential. To review the viral vector-based gene therapy landscape, we focus on these three highly regarded vector platforms and describe mechanisms of action and their roles in treating human disease.
Conflict of interest statement
G.G. is a scientific co-founder of Voyager Therapeutics, Adrenas Therapeutics, and Aspa Therapeutics, and holds equity in these companies. G.G. is an inventor on patents with potential royalties licensed to Voyager Therapeutics, Aspa Therapeutics, and other biopharmaceutical companies. The remaining authors declare no competing interests.
Figures



References
-
- Kohn DB, et al. Establishment and characterization of adenosine deaminase-deficient human T cell lines. J. Immunol. 1989;142:3971–3977. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

