Epidemiology and evolution of novel deltacoronaviruses in birds in central China
- PMID: 33559368
- PMCID: PMC8014545
- DOI: 10.1111/tbed.14029
Epidemiology and evolution of novel deltacoronaviruses in birds in central China
Abstract
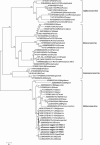
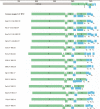
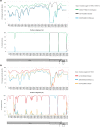
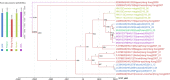
The variety and widespread of coronavirus in natural reservoir animals is likely to cause epidemics via interspecific transmission, which has attracted much attention due to frequent coronavirus epidemics in recent decades. Birds are natural reservoir of various viruses, but the existence of coronaviruses in wild birds in central China has been barely studied. Some bird coronaviruses belong to the genus of Deltacoronavirus. To explore the diversity of bird deltacoronaviruses in central China, we tested faecal samples from 415 wild birds in Hunan Province, China. By RT-PCR detection, we identified eight samples positive for deltacoronaviruses which were all from common magpies, and in four of them, we successfully amplified complete deltacoronavirus genomes distinct from currently known deltacoronavirus, indicating four novel deltacoronavirus stains (HNU1-1, HNU1-2, HNU2 and HNU3). Comparative analysis on the four genomic sequences showed that these novel magpie deltacoronaviruses shared three different S genes among which the S genes of HNU1-1 and HNU1-2 showed 93.8% amino acid (aa) identity to that of thrush coronavirus HKU12, HNU2 S showed 71.9% aa identity to that of White-eye coronavirus HKU16, and HNU3 S showed 72.4% aa identity to that of sparrow coronavirus HKU17. Recombination analysis showed that frequent recombination events of the S genes occurred among these deltacoronavirus strains. Two novel putative cleavage sites separating the non-structural proteins in the HNU coronaviruses were found. Bayesian phylogeographic analysis showed that the south coast of China might be a potential origin of bird deltacoronaviruses existing in inland China. In summary, these results suggest that common magpie in China carries diverse deltacoronaviruses with novel genomic features, indicating an important source of environmental coronaviruses closed to human communities, which may provide key information for prevention and control of future coronavirus epidemics.
Keywords: bird deltacoronavirus; common magpie; genome recombination; spike; viral genome.
© 2021 Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Discovery and Sequence Analysis of Four Deltacoronaviruses from Birds in the Middle East Reveal Interspecies Jumping with Recombination as a Potential Mechanism for Avian-to-Avian and Avian-to-Mammalian Transmission.J Virol. 2018 Jul 17;92(15):e00265-18. doi: 10.1128/JVI.00265-18. Print 2018 Aug 1. J Virol. 2018. PMID: 29769348 Free PMC article.
-
Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus.J Virol. 2012 Apr;86(7):3995-4008. doi: 10.1128/JVI.06540-11. Epub 2012 Jan 25. J Virol. 2012. PMID: 22278237 Free PMC article.
-
Diversity of gammacoronaviruses and deltacoronaviruses in wild birds and poultry in Russia.Sci Rep. 2022 Nov 12;12(1):19412. doi: 10.1038/s41598-022-23925-z. Sci Rep. 2022. PMID: 36371465 Free PMC article.
-
Wild birds as reservoirs for diverse and abundant gamma- and deltacoronaviruses.FEMS Microbiol Rev. 2020 Sep 1;44(5):631-644. doi: 10.1093/femsre/fuaa026. FEMS Microbiol Rev. 2020. PMID: 32672814 Free PMC article. Review.
-
[Research Advances in the Porcine Deltacoronavirus].Bing Du Xue Bao. 2016 Mar;32(2):243-8. Bing Du Xue Bao. 2016. PMID: 27396171 Review. Chinese.
Cited by
-
An Overview of the Importance and Value of Porcine Species in Sialic Acid Research.Biology (Basel). 2022 Jun 11;11(6):903. doi: 10.3390/biology11060903. Biology (Basel). 2022. PMID: 35741423 Free PMC article. Review.
-
High throughput screening for human disease associated-pathogens and antimicrobial resistance genes in migratory birds at ten habitat sites in China.BMC Microbiol. 2025 Jun 6;25(1):355. doi: 10.1186/s12866-025-04059-4. BMC Microbiol. 2025. PMID: 40481427 Free PMC article.
-
The taxonomy, host range and pathogenicity of coronaviruses and other viruses in the Nidovirales order.Anim Dis. 2021;1(1):5. doi: 10.1186/s44149-021-00005-9. Epub 2021 Apr 23. Anim Dis. 2021. PMID: 34778878 Free PMC article. Review.
-
Characterization of Deltacoronavirus in Black-Headed Gulls (Chroicocephalus ridibundus) in South China Indicating Frequent Interspecies Transmission of the Virus in Birds.Front Microbiol. 2022 May 12;13:895741. doi: 10.3389/fmicb.2022.895741. eCollection 2022. Front Microbiol. 2022. PMID: 35633699 Free PMC article.
-
Metagenomics of gut microbiome for migratory seagulls in Kunming city revealed the potential public risk to human health.BMC Genomics. 2023 May 19;24(1):269. doi: 10.1186/s12864-023-09379-1. BMC Genomics. 2023. PMID: 37208617 Free PMC article.
References
-
- Baele, G. , Lemey, P. , Bedford, T. , Rambaut, A. , Suchard, M. A. , & Alekseyenko, A. V. (2012). Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Molecular Biology and Evolution, 29(9), 2157–2167. 10.1093/molbev/mss084 - DOI - PMC - PubMed
-
- Chen, Q. I. , Wang, L. , Yang, C. , Zheng, Y. , Gauger, P. C. , Anderson, T. , Harmon, K. M. , Zhang, J. , Yoon, K.‐J. , Main, R. G. , & Li, G. (2018). The emergence of novel sparrow deltacoronaviruses in the United States more closely related to porcine deltacoronaviruses than sparrow deltacoronavirus HKU17. Emerging Microbes & Infections, 7(1), 105. 10.1038/s41426-018-0108-z - DOI - PMC - PubMed
MeSH terms
Grants and funding
- 2017YFD0500104/National Key Research and Development Program of China
- 32041001/National Natural Science Foundation of China
- 81902070/National Natural Science Foundation of China
- 2019JJ20004/Provincial Natural Science Foundation of Hunan Province
- 2019JJ50035/Provincial Natural Science Foundation of Hunan Province
LinkOut - more resources
Full Text Sources
Other Literature Sources

