Repurposing potential of Ayurvedic medicinal plants derived active principles against SARS-CoV-2 associated target proteins revealed by molecular docking, molecular dynamics and MM-PBSA studies
- PMID: 33561649
- PMCID: PMC7857054
- DOI: 10.1016/j.biopha.2021.111356
Repurposing potential of Ayurvedic medicinal plants derived active principles against SARS-CoV-2 associated target proteins revealed by molecular docking, molecular dynamics and MM-PBSA studies
Abstract
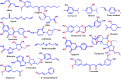
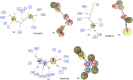
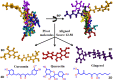
All the plants and their secondary metabolites used in the present study were obtained from Ayurveda, with historical roots in the Indian subcontinent. The selected secondary metabolites have been experimentally validated and reported as potent antiviral agents against genetically-close human viruses. The plants have also been used as a folk medicine to treat cold, cough, asthma, bronchitis, and severe acute respiratory syndrome in India and across the globe since time immemorial. The present study aimed to assess the repurposing possibility of potent antiviral compounds with SARS-CoV-2 target proteins and also with host-specific receptor and activator protease that facilitates the viral entry into the host body. Molecular docking (MDc) was performed to study molecular affinities of antiviral compounds with aforesaid target proteins. The top-scoring conformations identified through docking analysis were further validated by 100 ns molecular dynamic (MD) simulation run. The stability of the conformation was studied in detail by investigating the binding free energy using MM-PBSA method. Finally, the binding affinities of all the compounds were also compared with a reference ligand, remdesivir, against the target protein RdRp. Additionally, pharmacophore features, 3D structure alignment of potent compounds and Bayesian machine learning model were also used to support the MDc and MD simulation. Overall, the study emphasized that curcumin possesses a strong binding ability with host-specific receptors, furin and ACE2. In contrast, gingerol has shown strong interactions with spike protein, and RdRp and quercetin with main protease (Mpro) of SARS-CoV-2. In fact, all these target proteins play an essential role in mediating viral replication, and therefore, compounds targeting aforesaid target proteins are expected to block the viral replication and transcription. Overall, gingerol, curcumin and quercetin own multitarget binding ability that can be used alone or in combination to enhance therapeutic efficacy against COVID-19. The obtained results encourage further in vitro and in vivo investigations and also support the traditional use of antiviral plants preventively.
Keywords: ACE-2; COVID-19; Coronavirus; Furin; Main protease; RdRp, spike protein RBD.
Copyright © 2021. Published by Elsevier Masson SAS.
Conflict of interest statement
There is no conflict of interest for the publication of this article.
Figures









References
-
- W.H. Organization, Coronavirus Disease 2019 (COVID-19): Situation Report, 67, 2020, pp. 1–18.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

