Epigenetics: New Insights into Mammary Gland Biology
- PMID: 33562534
- PMCID: PMC7914701
- DOI: 10.3390/genes12020231
Epigenetics: New Insights into Mammary Gland Biology
Abstract
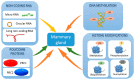
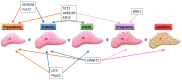
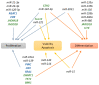
The mammary gland undergoes important anatomical and physiological changes from embryogenesis through puberty, pregnancy, lactation and involution. These steps are under the control of a complex network of molecular factors, in which epigenetic mechanisms play a role that is increasingly well described. Recently, studies investigating epigenetic modifications and their impacts on gene expression in the mammary gland have been performed at different physiological stages and in different mammary cell types. This has led to the establishment of a role for epigenetic marks in milk component biosynthesis. This review aims to summarize the available knowledge regarding the involvement of the four main molecular mechanisms in epigenetics: DNA methylation, histone modifications, polycomb protein activity and non-coding RNA functions.
Keywords: DNA methylation; epigenetic regulations; lactation; mammary gland; non-coding RNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

