Segregation of brain and organizer precursors is differentially regulated by Nodal signaling at blastula stage
- PMID: 33563608
- PMCID: PMC7928228
- DOI: 10.1242/bio.051797
Segregation of brain and organizer precursors is differentially regulated by Nodal signaling at blastula stage
Abstract
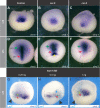
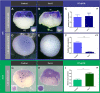
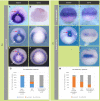
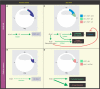
The blastula Chordin- and Noggin-expressing (BCNE) center comprises animal-dorsal and marginal-dorsal cells of the amphibian blastula and contains the precursors of the brain and the gastrula organizer. Previous findings suggested that the BCNE behaves as a homogeneous cell population that only depends on nuclear β-catenin activity but does not require Nodal and later segregates into its descendants during gastrulation. In contrast to previous findings, in this work, we show that the BCNE does not behave as a homogeneous cell population in response to Nodal antagonists. In fact, we found that chordin.1 expression in a marginal subpopulation of notochordal precursors indeed requires Nodal input. We also establish that an animal BCNE subpopulation of cells that express both, chordin.1 and sox2 (a marker of pluripotent neuroectodermal cells), and gives rise to most of the brain, persisted at blastula stage after blocking Nodal. Therefore, Nodal signaling is required to define a population of chordin.1+ cells and to restrict the recruitment of brain precursors within the BCNE as early as at blastula stage. We discuss our findings in Xenopus in comparison to other vertebrate models, uncovering similitudes in early brain induction and delimitation through Nodal signaling.
Keywords: BCNE center; Brain; Chordin; Gastrula organizer; Nodal; Vertebrates.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures



References
-
- Anderson, R. M., Lawrence, A. R., Stottmann, R. W., Bachiller, D. and Klingensmith, J. (2002). Chordin and noggin promote organizing centers of forebrain development in the mouse. Development 129, 4975-4987. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

