Matched Targeted Therapy for Pediatric Patients with Relapsed, Refractory, or High-Risk Leukemias: A Report from the LEAP Consortium
- PMID: 33563661
- PMCID: PMC8178162
- DOI: 10.1158/2159-8290.CD-20-0564
Matched Targeted Therapy for Pediatric Patients with Relapsed, Refractory, or High-Risk Leukemias: A Report from the LEAP Consortium
Abstract
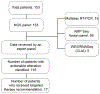
Despite a remarkable increase in the genomic profiling of cancer, integration of genomic discoveries into clinical care has lagged behind. We report the feasibility of rapid identification of targetable mutations in 153 pediatric patients with relapsed/refractory or high-risk leukemias enrolled on a prospective clinical trial conducted by the LEAP Consortium. Eighteen percent of patients had a high confidence Tier 1 or 2 recommendation. We describe clinical responses in the 14% of patients with relapsed/refractory leukemia who received the matched targeted therapy. Further, in order to inform future targeted therapy for patients, we validated variants of uncertain significance, performed ex vivo drug-sensitivity testing in patient leukemia samples, and identified new combinations of targeted therapies in cell lines and patient-derived xenograft models. These data and our collaborative approach should inform the design of future precision medicine trials. SIGNIFICANCE: Patients with relapsed/refractory leukemias face limited treatment options. Systematic integration of precision medicine efforts can inform therapy. We report the feasibility of identifying targetable mutations in children with leukemia and describe correlative biology studies validating therapeutic hypotheses and novel mutations.See related commentary by Bornhauser and Bourquin, p. 1322.This article is highlighted in the In This Issue feature, p. 1307.
©2021 American Association for Cancer Research.
Figures



Comment in
-
A Hopeful Leap Forward by Multicentric Cooperation for Precision-Based Therapy for Very Resistant, Relapsed, or Refractory Childhood Leukemia.Cancer Discov. 2021 Jun;11(6):1322-1323. doi: 10.1158/2159-8290.CD-21-0258. Cancer Discov. 2021. PMID: 34078660
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

