Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia
- PMID: 33563978
- PMCID: PMC7873279
- DOI: 10.1038/s41467-021-21240-1
Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia
Erratum in
-
Author Correction: Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia.Nat Commun. 2021 Feb 25;12(1):1430. doi: 10.1038/s41467-021-21768-2. Nat Commun. 2021. PMID: 33633118 Free PMC article. No abstract available.
Abstract
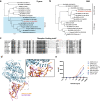
Among the many questions unanswered for the COVID-19 pandemic are the origin of SARS-CoV-2 and the potential role of intermediate animal host(s) in the early animal-to-human transmission. The discovery of RaTG13 bat coronavirus in China suggested a high probability of a bat origin. Here we report molecular and serological evidence of SARS-CoV-2 related coronaviruses (SC2r-CoVs) actively circulating in bats in Southeast Asia. Whole genome sequences were obtained from five independent bats (Rhinolophus acuminatus) in a Thai cave yielding a single isolate (named RacCS203) which is most related to the RmYN02 isolate found in Rhinolophus malayanus in Yunnan, China. SARS-CoV-2 neutralizing antibodies were also detected in bats of the same colony and in a pangolin at a wildlife checkpoint in Southern Thailand. Antisera raised against the receptor binding domain (RBD) of RmYN02 was able to cross-neutralize SARS-CoV-2 despite the fact that the RBD of RacCS203 or RmYN02 failed to bind ACE2. Although the origin of the virus remains unresolved, our study extended the geographic distribution of genetically diverse SC2r-CoVs from Japan and China to Thailand over a 4800-km range. Cross-border surveillance is urgently needed to find the immediate progenitor virus of SARS-CoV-2.
Conflict of interest statement
L.-F.W., C.W.T., and W.N.C. are inventors of a patent application for the SARS-CoV-2 sVNT, which has been commercialized by GenScript Biotech under the tradename cPassTM. Other authors declare no competing interests.
Figures



References
-
- WHO. COVID-19 status report. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situatio... (2020).
-
- Callaway, E. & Cyranoski, D. China coronavirus: six questions scientists are asking. Nature577, 605–607 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

