19F NMR as a tool in chemical biology
- PMID: 33564338
- PMCID: PMC7849273
- DOI: 10.3762/bjoc.17.28
19F NMR as a tool in chemical biology
Abstract
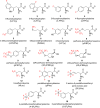
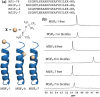
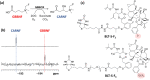
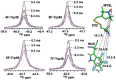
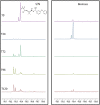
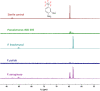
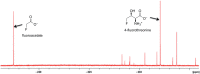
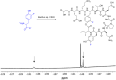
We previously reviewed the use of 19F NMR in the broad field of chemical biology [Cobb, S. L.; Murphy, C. D. J. Fluorine Chem. 2009, 130, 132-140] and present here a summary of the literature from the last decade that has the technique as the central method of analysis. The topics covered include the synthesis of new fluorinated probes and their incorporation into macromolecules, the application of 19F NMR to monitor protein-protein interactions, protein-ligand interactions, physiologically relevant ions and in the structural analysis of proteins and nucleic acids. The continued relevance of the technique to investigate biosynthesis and biodegradation of fluorinated organic compounds is also described.
Keywords: 19F NMR; biotransformation; chemical biology; fluorine; probes; protein structure.
Copyright © 2021, Gimenez et al.
Figures













References
-
- Cobb S L, Murphy C D. J Fluorine Chem. 2009;130:132–143. doi: 10.1016/j.jfluchem.2008.11.003. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
