This is a preprint.
A Molecular network approach reveals shared cellular and molecular signatures between chronic fatigue syndrome and other fatiguing illnesses
- PMID: 33564792
- PMCID: PMC7872387
- DOI: 10.1101/2021.01.29.21250755
A Molecular network approach reveals shared cellular and molecular signatures between chronic fatigue syndrome and other fatiguing illnesses
Conflict of interest statement
Competing interest statement: The external patient data used to build the Bayesian network were partially funded as part of research alliance between Janssen Biotech and The Icahn School of Medicine at Mount Sinai.
Figures
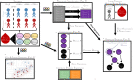
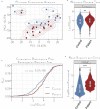


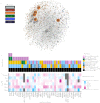
References
-
- Greenhalgh T., et al. , Management of post-acute covid-19 in primary care. BMJ, 2020. 370: p. m3026. - PubMed
-
- Rubin R., As Their Numbers Grow, COVID-19 “Long Haulers” Stump Experts. JAMA, 2020. 324(14): p. 1381–1383. - PubMed
-
- CDC. [Website] 2018; Available from: https://www.cdc.gov/me-cfs/about/possiblecauses.html.
-
- Tom Whipple O.M., Scientists trade insults over myalgic encephalomyelitis (ME) study. The Times, 2017.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
