Perinatal granulopoiesis and risk of pediatric asthma
- PMID: 33565964
- PMCID: PMC7889076
- DOI: 10.7554/eLife.63745
Perinatal granulopoiesis and risk of pediatric asthma
Abstract
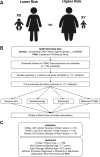
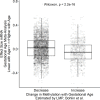
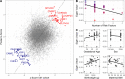
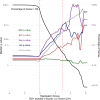
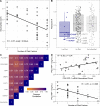
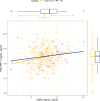
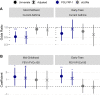
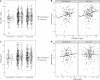
There are perinatal characteristics, such as gestational age, reproducibly associated with the risk for pediatric asthma. Identification of biologic processes influenced by these characteristics could facilitate risk stratification or new therapeutic targets. We hypothesized that transcriptional changes associated with multiple epidemiologic risk factors would be mediators of pediatric asthma risk. Using publicly available transcriptomic data from cord blood mononuclear cells, transcription of genes involved in myeloid differentiation was observed to be inversely associated with a pediatric asthma risk stratification based on multiple perinatal risk factors. This gene signature was validated in an independent prospective cohort and was specifically associated with genes localizing to neutrophil-specific granules. Further validation demonstrated that umbilical cord blood serum concentration of PGLYRP-1, a specific granule protein, was inversely associated with mid-childhood current asthma and early-teen FEV1/FVCx100. Thus, neutrophil-specific granule abundance at birth predicts risk for pediatric asthma and pulmonary function in adolescence.
Trial registration: ClinicalTrials.gov NCT02820402.
Keywords: allergy; asthma; fetal blood; granulocytes; human; immunology; inflammation; medicine; peptidoglycan recognition protein; risk factor.
© 2021, Turturice et al.
Conflict of interest statement
BT, JT, MK, LT, DG, AL, EO, SR, DP, PF No competing interests declared
Figures



References
-
- Azad MB, Coneys JG, Kozyrskyj AL, Field CJ, Ramsey CD, Becker AB, Friesen C, Abou-Setta AM, Zarychanski R. Probiotic supplementation during pregnancy or infancy for the prevention of asthma and wheeze: systematic review and meta-analysis. BMJ. 2013;347:f6471. doi: 10.1136/bmj.f6471. - DOI - PMC - PubMed
-
- Bui DS, Lodge CJ, Burgess JA, Lowe AJ, Perret J, Bui MQ, Bowatte G, Gurrin L, Johns DP, Thompson BR, Hamilton GS, Frith PA, James AL, Thomas PS, Jarvis D, Svanes C, Russell M, Morrison SC, Feather I, Allen KJ, Wood-Baker R, Hopper J, Giles GG, Abramson MJ, Walters EH, Matheson MC, Dharmage SC. Childhood predictors of lung function trajectories and future COPD risk: a prospective cohort study from the first to the sixth decade of life. The Lancet Respiratory Medicine. 2018;6:535–544. doi: 10.1016/S2213-2600(18)30100-0. - DOI - PubMed
-
- Bukowski R, Sadovsky Y, Goodarzi H, Zhang H, Biggio JR, Varner M, Parry S, Xiao F, Esplin SM, Andrews W, Saade GR, Ilekis JV, Reddy UM, Baldwin DA. Onset of human preterm and term birth is related to unique inflammatory transcriptome profiles at the maternal fetal interface. PeerJ. 2017;5:e3685. doi: 10.7717/peerj.3685. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

