Revisiting soil bacterial counting methods: Optimal soil storage and pretreatment methods and comparison of culture-dependent and -independent methods
- PMID: 33566842
- PMCID: PMC7875414
- DOI: 10.1371/journal.pone.0246142
Revisiting soil bacterial counting methods: Optimal soil storage and pretreatment methods and comparison of culture-dependent and -independent methods
Abstract
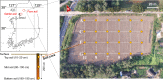
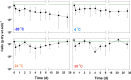
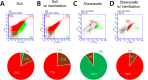
Although a number of different methods have been used to quantify soil bacteria, identifying the optimal method(s) for soil bacterial abundance is still in question. No single method exists for undertaking an absolute microbial count using culture-dependent methods (CDMs) or even culture-independent methods (CIMs). This study investigated soil storage and pretreatment methods for optimal bacterial counts. Appropriate storage temperature (4°C) and optimal pretreatment methods (sonication time for 3 min and centrifugation at 1400 g) were necessary to preserve bacterial cell viability and eliminate interference from soil particles. To better estimate soil bacterial numbers under various cellular state and respiration, this study also evaluated three CDMs (i.e., colony forming unit, spotting, and most probable number (MPN) and three CIMs (i.e., flow cytometry (FCM), epifluorescence microscopy (EM) count, and DNA quantitation). Each counting method was tested using 72 soil samples collected from a local arable farm site at three different depths (i.e., 10-20, 90-100, and 180-190 cm). Among all CDMs, MPN was found to be rapid, simple, and reliable. However, the number of bacteria quantified by MPN was 1-2 orders lower than that quantified by CIMs, likely due to the inability of MPN to count anaerobic bacteria. The DNA quantitation method appeared to overestimate soil bacterial numbers, which may be attributed to DNA from dead bacteria and free DNA in the soil matrix. FCM was found to be ineffective in counting soil bacteria as it was difficult to separate the bacterial cells from the soil particles. Dyes used in FCM stained the bacterial DNA and clay particles. The EM count was deemed a highly effective method as it provided information on soil mineral particles, live bacteria, and dead bacteria; however, it was a time-consuming and labor-intensive process. Combining both types of methods was considered the best approach to acquire better information on the characteristics of indigenous soil microorganisms (aerobic versus anaerobic, live versus dead).
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- BÁRcenas-Moreno G, GÓMez-BrandÓN M, Rousk J, BÅÅTh E. Adaptation of soil microbial communities to temperature: comparison of fungi and bacteria in a laboratory experiment. Glob Change Biol. 2009;15(12):2950–7. 10.1111/j.1365-2486.2009.01882.x - DOI
-
- Curd EE, Martiny JBH, Li H, Smith TB. Bacterial diversity is positively correlated with soil heterogeneity. Ecosphere. 2018;9(1):e02079 10.1002/ecs2.2079 - DOI
-
- Tuzimura K, Watanabe I. Estimation of number of root-nodule bacteria by a nodulation-dilution frequency method. Soil Sci Plant Nutr. 1961;7(2):61–5. 10.1080/00380768.1961.10430958 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

