MiR396-GRF module associates with switchgrass biomass yield and feedstock quality
- PMID: 33567151
- PMCID: PMC8384596
- DOI: 10.1111/pbi.13567
MiR396-GRF module associates with switchgrass biomass yield and feedstock quality
Abstract
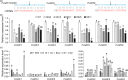
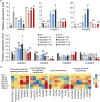
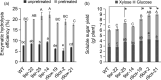
Improving plant biomass yield and/or feedstock quality for highly efficient lignocellulose conversion has been the main research focus in genetic modification of switchgrass (Panicum virgatum L.), a dedicated model plant for biofuel production. Here, we proved that overexpression of miR396 (OE-miR396) leads to reduced plant height and lignin content mainly by reducing G-lignin monomer content. We identified nineteen PvGRFs in switchgrass and proved thirteen of them were cleaved by miR396. MiR396-targeted PvGRF1, PvGRF9 and PvGRF3 showed significantly higher expression in stem. By separately overexpressing rPvGRF1, 3 and 9, in which synonymous mutations abolished the miR396 target sites, and suppression of PvGRF1/3/9 activity via PvGRF1/3/9-SRDX overexpression in switchgrass, we confirmed PvGRF1 and PvGRF9 played positive roles in improving plant height and G-lignin content. Overexpression of PvGRF9 was sufficient to complement the defective phenotype of OE-miR396 plants. MiR396-PvGRF9 modulates these traits partly by interfering GA and auxin biosynthesis and signalling transduction and cell wall lignin, glucose and xylan biosynthesis pathways. Moreover, by enzymatic hydrolysis analyses, we found that overexpression of rPvGRF9 significantly enhanced per plant sugar yield. Our results suggest that PvGRF9 can be utilized as a candidate molecular tool in modifying plant biomass yield and feedstock quality.
Keywords: GRF; biomass yield; feedstock quality; miR396; switchgrass.
© 2021 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interests.
Figures




References
-
- Cao, D. , Wang, J. , Ju, Z. , Liu, Q. , Li, S. , Tian, H. , Fu, D. , Zhu, H. , Luo, Y. and Zhu, B. (2016) Regulations on growth and development in tomato cotyledon, flower and fruit via destruction of miR396 with short tandem target mimic. Plant Sci. 247, 1–12. - PubMed
-
- Chabannes, M. , Barakate, A. , Lapierre, C. , Marita, J.M. , Ralph, J. , Pean, M. and Danoun, S. (2001) Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down‐regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 28, 257–270. - PubMed
-
- Chandran, V. , Wang, H. , Gao, F. , Cao, X.L. , Chen, Y.P. , Li, G.B. , Zhu, Y. , Yang, X.M. , Zhang, L.L. , Zhao, Z.X. , Zhao, J.H. , Wang, Y.G. , Li, S. , Fan, J. , Li, Y. , Zhao, J.Q. , Li, S.Q. and Wang, W.M. (2019) miR396‐OsGRFs module balances growth and rice blast disease‐resistance. Frontiers Plant Sci. 9, 1999. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

