Structure and mechanism of a phage-encoded SAM lyase revises catalytic function of enzyme family
- PMID: 33567250
- PMCID: PMC7877911
- DOI: 10.7554/eLife.61818
Structure and mechanism of a phage-encoded SAM lyase revises catalytic function of enzyme family
Abstract
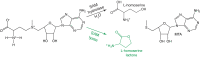
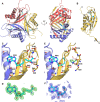
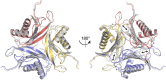
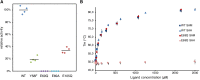
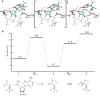
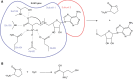
The first S-adenosyl methionine (SAM) degrading enzyme (SAMase) was discovered in bacteriophage T3, as a counter-defense against the bacterial restriction-modification system, and annotated as a SAM hydrolase forming 5'-methyl-thioadenosine (MTA) and L-homoserine. From environmental phages, we recently discovered three SAMases with barely detectable sequence similarity to T3 SAMase and without homology to proteins of known structure. Here, we present the very first phage SAMase structures, in complex with a substrate analogue and the product MTA. The structure shows a trimer of alpha-beta sandwiches similar to the GlnB-like superfamily, with active sites formed at the trimer interfaces. Quantum-mechanical calculations, thin-layer chromatography, and nuclear magnetic resonance spectroscopy demonstrate that this family of enzymes are not hydrolases but lyases forming MTA and L-homoserine lactone in a unimolecular reaction mechanism. Sequence analysis and in vitro and in vivo mutagenesis support that T3 SAMase belongs to the same structural family and utilizes the same reaction mechanism.
Keywords: E. coli; S-adenosyl methionine; bacteriophage; biochemistry; chemical biology; lyase; molecular biophysics; structural biology.
Plain language summary
Bacteria can be infected by viruses known as bacteriophages. These viruses inject their genetic material into bacterial cells and use the bacteria’s own machinery to build the proteins they need to survive and infect other cells. To protect themselves, bacteria produce a molecule called S-adenosyl methionine, or SAM for short, which deposits marks on the bacteria’s DNA. These marks help the bacteria distinguish their own genetic material from the genetic material of foreign invaders: any DNA not bearing the mark from SAM will be immediately broken down by the bacterial cell. This system helps to block many types of bacteriophage infections, but not all. Some bacteriophages carry genes that code for enzymes called SAMases, which can break down SAM, switching off the bacteria’s defenses. The most well-known SAMase was first discovered in the 1960s in a bacteriophage called T3. Chemical studies of this SAMase suggested that it works as a 'hydrolase', meaning that it uses water to break SAM apart. New SAMases have since been discovered in bacteriophages from environmental water samples, which, despite being able to degrade SAM, are genetically dissimilar to one another and the SAMase in T3. This brings into question whether these enzymes all use the same mechanism to break SAM down. To gain a better understanding of how these SAMases work, Guo, Söderholm, Kanchugal, Isaksen et al. solved the crystal structure of one of the newly discovered enzymes called Svi3-3. This revealed three copies of the Svi3-3 enzyme join together to form a unit that SAM binds to at the border between two of the enzymes. Computer simulations of this structure suggested that Svi3-3 holds SAM in a position where it cannot interact with water, and that once in the grip of the SAMase, SAM instead reacts with itself and splits into two. Experiments confirmed these predictions for Svi3-3 and the other tested SAMases. Furthermore, the SAMase from bacteriophage T3 was also found to degrade SAM using the same mechanism. This shows that this group of SAMases are not hydrolases as originally thought, but in fact ‘lyases’: enzymes that break molecules apart without using water. These findings form a starting point for further investigations into how SAM lyases help bacteriophages evade detection. SAM has various different functions in other living organisms, and these lyases could be used to modulate the levels of SAM in future studies investigating its role.
© 2021, Guo et al.
Conflict of interest statement
XG, AS, SK, GI, OW, UE, ST, AG, JJ, JÅ, DA, MS No competing interests declared
Figures









References
-
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica Section D Biological Crystallography. 2012;68:352–367. doi: 10.1107/S0907444912001308. - DOI - PMC - PubMed
-
- Becke AD. Density‐functional thermochemistry. III. the role of exact exchange. The Journal of Chemical Physics. 1993;98:5648–5652. doi: 10.1063/1.464913. - DOI
-
- Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, Salmon JK, Shan Y, Shaw DE. Scalable algorithms for molecular dynamics simulations on commodity clusters. Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, SC; 2006. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

