COVID-19 immune signatures reveal stable antiviral T cell function despite declining humoral responses
- PMID: 33567252
- PMCID: PMC7871825
- DOI: 10.1016/j.immuni.2021.01.008
COVID-19 immune signatures reveal stable antiviral T cell function despite declining humoral responses
Abstract
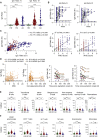
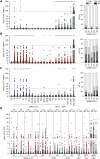
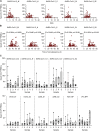
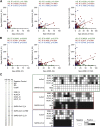
Cellular and humoral immunity to SARS-CoV-2 is critical to control primary infection and correlates with severity of disease. The role of SARS-CoV-2-specific T cell immunity, its relationship to antibodies, and pre-existing immunity against endemic coronaviruses (huCoV), which has been hypothesized to be protective, were investigated in 82 healthy donors (HDs), 204 recovered (RCs), and 92 active COVID-19 patients (ACs). ACs had high amounts of anti-SARS-CoV-2 nucleocapsid and spike IgG but lymphopenia and overall reduced antiviral T cell responses due to the inflammatory milieu, expression of inhibitory molecules (PD-1, Tim-3) as well as effector caspase-3, -7, and -8 activity in T cells. SARS-CoV-2-specific T cell immunity conferred by polyfunctional, mainly interferon-γ-secreting CD4+ T cells remained stable throughout convalescence, whereas humoral responses declined. Immune responses toward huCoV in RCs with mild disease and strong cellular SARS-CoV-2 T cell reactivity imply a protective role of pre-existing immunity against huCoV.
Keywords: COVID-19; SARS-CoV-2; antibody; antiviral T cell immunity; caspases; cell death; chemokine receptors; convalescence; endemic human coronavirus; humoral immunity.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare that they have no competing interests. The funders played no role in designing the study, in collecting, analyzing, or interpreting the data, in writing the manuscript, or in the decision to publish the results.
Figures



References
-
- Behrens G.M.N., Cossmann A., Stankov M.V., Schulte B., Streeck H., Förster R., Bosnjak B., Willenzon S., Boeck A.L., Thu Tran A., et al. Strategic Anti-SARS-CoV-2 Serology Testing in a Low Prevalence Setting: The COVID-19 Contact (CoCo) Study in Healthcare Professionals. Infect. Dis. Ther. 2020;9:837–849. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

