SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO
- PMID: 33567255
- PMCID: PMC7857071
- DOI: 10.1016/j.celrep.2021.108761
SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO
Abstract
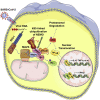
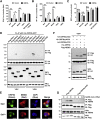
Coronavirus disease 2019 (COVID-19) is a current global health threat caused by the novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Emerging evidence indicates that SARS-CoV-2 elicits a dysregulated immune response and a delayed interferon (IFN) expression in patients, which contribute largely to the viral pathogenesis and development of COVID-19. However, underlying mechanisms remain to be elucidated. Here, we report the activation and repression of the innate immune response by SARS-CoV-2. We show that SARS-CoV-2 RNA activates the RIG-I-MAVS-dependent IFN signaling pathway. We further uncover that ORF9b immediately accumulates and antagonizes the antiviral type I IFN response during SARS-CoV-2 infection on primary human pulmonary alveolar epithelial cells. ORF9b targets the nuclear factor κB (NF-κB) essential modulator NEMO and interrupts its K63-linked polyubiquitination upon viral stimulation, thereby inhibiting the canonical IκB kinase alpha (IKKα)/β/γ-NF-κB signaling and subsequent IFN production. Our findings thus unveil the innate immunosuppression by ORF9b and provide insights into the host-virus interplay during the early stage of SARS-CoV-2 infection.
Keywords: COVID-19; RIG-I-MAVS signaling; SARS-CoV-2; innate immune response; interferon.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Bhoj V.G., Chen Z.J. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

