mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants
- PMID: 33567448
- PMCID: PMC8503938
- DOI: 10.1038/s41586-021-03324-6
mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants
Abstract
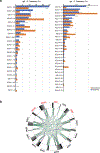
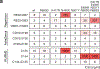
Here we report on the antibody and memory B cell responses of a cohort of 20 volunteers who received the Moderna (mRNA-1273) or Pfizer-BioNTech (BNT162b2) vaccine against SARS-CoV-21-4. Eight weeks after the second injection of vaccine, volunteers showed high levels of IgM and IgG anti-SARS-CoV-2 spike protein (S) and receptor-binding-domain (RBD) binding titre. Moreover, the plasma neutralizing activity and relative numbers of RBD-specific memory B cells of vaccinated volunteers were equivalent to those of individuals who had recovered from natural infection5,6. However, activity against SARS-CoV-2 variants that encode E484K-, N501Y- or K417N/E484K/N501-mutant S was reduced by a small-but significant-margin. The monoclonal antibodies elicited by the vaccines potently neutralize SARS-CoV-2, and target a number of different RBD epitopes in common with monoclonal antibodies isolated from infected donors5-8. However, neutralization by 14 of the 17 most-potent monoclonal antibodies that we tested was reduced or abolished by the K417N, E484K or N501Y mutation. Notably, these mutations were selected when we cultured recombinant vesicular stomatitis virus expressing SARS-CoV-2 S in the presence of the monoclonal antibodies elicited by the vaccines. Together, these results suggest that the monoclonal antibodies in clinical use should be tested against newly arising variants, and that mRNA vaccines may need to be updated periodically to avoid a potential loss of clinical efficacy.
Conflict of interest statement
Figures
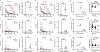
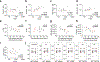








Update of
-
mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants.bioRxiv [Preprint]. 2021 Jan 30:2021.01.15.426911. doi: 10.1101/2021.01.15.426911. bioRxiv. 2021. Update in: Nature. 2021 Apr;592(7855):616-622. doi: 10.1038/s41586-021-03324-6. PMID: 33501451 Free PMC article. Updated. Preprint.
Comment in
-
SARS-CoV-2 mutations, vaccines, and immunity: implication of variants of concern.Signal Transduct Target Ther. 2021 May 22;6(1):203. doi: 10.1038/s41392-021-00623-2. Signal Transduct Target Ther. 2021. PMID: 34023862 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- P50 AI150464-13/NH/NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 AI078788/AI/NIAID NIH HHS/United States
- P01 AI138938/AI/NIAID NIH HHS/United States
- R01 AI78788/NH/NIH HHS/United States
- R37 AI064003/AI/NIAID NIH HHS/United States
- U19 AI111825/AI/NIAID NIH HHS/United States
- P50 AI150464/AI/NIAID NIH HHS/United States
- P01 AI138398-S1/NH/NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- UL1 TR001866/TR/NCATS NIH HHS/United States
- 2U19AI111825/NH/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

