Epigenetic signatures in cancer: proper controls, current challenges and the potential for clinical translation
- PMID: 33568205
- PMCID: PMC7874645
- DOI: 10.1186/s13073-021-00837-7
Epigenetic signatures in cancer: proper controls, current challenges and the potential for clinical translation
Abstract
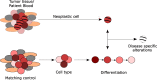
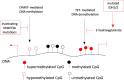
Epigenetic alterations are associated with normal biological processes such as aging or differentiation. Changes in global epigenetic signatures, together with genetic alterations, are driving events in several diseases including cancer. Comparative studies of cancer and healthy tissues found alterations in patterns of DNA methylation, histone posttranslational modifications, and changes in chromatin accessibility. Driven by sophisticated, next-generation sequencing-based technologies, recent studies discovered cancer epigenomes to be dominated by epigenetic patterns already present in the cell-of-origin, which transformed into a neoplastic cell. Tumor-specific epigenetic changes therefore need to be redefined and factors influencing epigenetic patterns need to be studied to unmask truly disease-specific alterations. The underlying mechanisms inducing cancer-associated epigenetic alterations are poorly understood. Studies of mutated epigenetic modifiers, enzymes that write, read, or edit epigenetic patterns, or mutated chromatin components, for example oncohistones, help to provide functional insights on how cancer epigenomes arise. In this review, we highlight the importance and define challenges of proper control tissues and cell populations to exploit cancer epigenomes. We summarize recent advances describing mechanisms leading to epigenetic changes in tumorigenesis and briefly discuss advances in investigating their translational potential.
Keywords: Cancer; Cell-of-origin; Epigenetic signatures; Epigenetic therapy; Epigenomics; Oncohistones; Precision oncology; Tumor subclassification.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

