PTM-Shepherd: Analysis and Summarization of Post-Translational and Chemical Modifications From Open Search Results
- PMID: 33568339
- PMCID: PMC7950090
- DOI: 10.1074/mcp.TIR120.002216
PTM-Shepherd: Analysis and Summarization of Post-Translational and Chemical Modifications From Open Search Results
Abstract
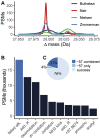
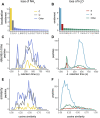
Open searching has proven to be an effective strategy for identifying both known and unknown modifications in shotgun proteomics experiments. Rather than being limited to a small set of user-specified modifications, open searches identify peptides with any mass shift that may correspond to a single modification or a combination of several modifications. Here we present PTM-Shepherd, a bioinformatics tool that automates characterization of post-translational modification profiles detected in open searches based on attributes, such as amino acid localization, fragmentation spectra similarity, retention time shifts, and relative modification rates. PTM-Shepherd can also perform multiexperiment comparisons for studying changes in modification profiles, e.g., in data generated in different laboratories or under different conditions. We demonstrate how PTM-Shepherd improves the analysis of data from formalin-fixed and paraffin-embedded samples, detects extreme underalkylation of cysteine in some data sets, discovers an artifactual modification introduced during peptide synthesis, and uncovers site-specific biases in sample preparation artifacts in a multicenter proteomics profiling study.
Keywords: Open searching, PTM, Post-translational modification, Mass-tolerant search, Localization, Spectral similarity, Retention time.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest Authors declare no competing interests.
Figures




References
-
- Nesvizhskii A.I., Roos F.F., Grossmann J., Vogelzang M., Eddes J.S., Gruissem W., Baginsky S., Aebersold R. Dynamic spectrum quality assessment and iterative computational analysis of shotgun proteomic data: toward more efficient identification of post-translational modifications, sequence polymorphisms, and novel peptides. Mol. Cell. Proteomics. 2006;5:652–670. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

