Clinical and Molecular Epidemiology of an Emerging Panton-Valentine Leukocidin-Positive ST5 Methicillin-Resistant Staphylococcus aureus Clone in Northern Australia
- PMID: 33568451
- PMCID: PMC8544886
- DOI: 10.1128/mSphere.00651-20
Clinical and Molecular Epidemiology of an Emerging Panton-Valentine Leukocidin-Positive ST5 Methicillin-Resistant Staphylococcus aureus Clone in Northern Australia
Abstract
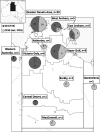
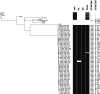
Recently, we identified a Staphylococcus aureus sequence type 5 (ST5) clone in northern Australia with discrepant trimethoprim-sulfamethoxazole (SXT) susceptibility results. We aimed to identify isolates of this clone using Vitek 2 SXT resistance as a proxy and to compare its epidemiology with those of other circulating S. aureus strains. We collated Vitek 2 susceptibility data for S. aureus isolates collected through our laboratory and conducted a prospective, case-control study comparing clinical, microbiological, epidemiological, and genomic data for subsets of isolates reported as SXT resistant (cases) and SXT susceptible (controls) by Vitek 2. While overall SXT resistance rates remained relatively stable from 2011 to 2018 among 27,721 S. aureus isolates, non-multidrug-resistant methicillin-resistant S. aureus (MRSA) strains almost completely replaced multidrug-resistant MRSA strains as the predominant SXT-resistant MRSA phenotype. Demographic and clinical features of 51 case-control pairs were similar, but genotyping revealed stark differences: clonal complex 5 (CC5) MRSA predominated among SXT-resistant cases (34/51 [67%]), while CC93 MRSA predominated among susceptible controls (26/51 [51%]). All CC5 isolates were an ST5 clonal lineage that possessed the trimethoprim resistance gene dfrG within SCCmec IVo; all were SXT susceptible by Etest. The replacement of Vitek 2 reported SXT-resistant multidrug-resistant MRSA by non-multidrug-resistant MRSA appears related to the emergence of an ST5-MRSA-SCCmec IVo clone that is SXT susceptible by Etest and causes clinical disease similar to that caused by ST93-MRSA-SCCmec IVa. Reliance on Vitek 2 SXT reporting may lead to unnecessary restriction of effective oral treatment options for S. aureus infections. Whether the presence of dfrG within SCCmec IVo provides a selective advantage at the population level is currently unclear.IMPORTANCEStaphylococcus aureus is an important human pathogen that causes a wide range of clinical infections. In the past 2 decades, an epidemic of community-associated skin and soft tissue infections has been driven by S. aureus strains with specific virulence factors and resistance to beta-lactam antibiotics. Recently, an S. aureus strain with discrepant antimicrobial susceptibility testing results has emerged in northern Australia. This ST5-MRSA-SCCmec IVo clone is reported as resistant to trimethoprim-sulfamethoxazole by Vitek 2 but susceptible by phenotypic methods. ST5-MRSA-SCCmec IVo is now the second most common community-associated MRSA clone in parts of Australia and causes a spectrum of clinical disease similar to that caused by the virulent ST93-MRSA lineage. Whole-genome sequence analysis demonstrates that ST5-MRSA-SCCmecIVo is causing a clonal outbreak across a large geographical region. Although phenotypic testing suggests in vitro susceptibility to trimethoprim-sulfamethoxazole, it is unclear at this stage whether the presence of dfrG within SCCmec IVo provides a selective advantage at the population level.
Keywords: Staphylococcus aureus; epidemiology; genomics; methicillin resistance; susceptibility testing; trimethoprim.
Copyright © 2021 McGuinness et al.
Figures
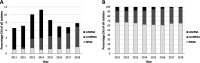


References
-
- Tong SY, Varrone L, Chatfield MD, Beaman M, Giffard PM. 2015. Progressive increase in community-associated methicillin-resistant Staphylococcus aureus in Indigenous populations in northern Australia from 1993 to 2012. Epidemiol Infect 143:1519–1523. doi: 10.1017/S0950268814002611. - DOI - PMC - PubMed
-
- Chua K, Laurent F, Coombs G, Grayson ML, Howden BP. 2011. Antimicrobial resistance: not community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA)! A clinician’s guide to community MRSA—its evolving antimicrobial resistance and implications for therapy. Clin Infect Dis 52:99–114. doi: 10.1093/cid/ciq067. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

