Deciphering an Adenovirus F41 Outbreak in Pediatric Hematopoietic Stem Cell Transplant Recipients by Whole-Genome Sequencing
- PMID: 33568462
- PMCID: PMC8091842
- DOI: 10.1128/JCM.03148-20
Deciphering an Adenovirus F41 Outbreak in Pediatric Hematopoietic Stem Cell Transplant Recipients by Whole-Genome Sequencing
Abstract
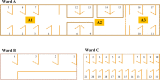
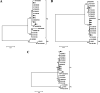
Human adenovirus (HAdV) represents a major cause of mortality and morbidity in pediatric recipients of allogeneic hematopoietic stem cell transplants (HSCT). HAdV species F type 41 (HAdV-F41) infections in HSCT patients are scarce, whereas HAdV-F41 circulates commonly in healthy individuals. Between March and July 2018, HAdV-F41 infections were identified in four children (A, B, C, and E) who received allogeneic HSCT and one child before HSCT (D) at Robert Debré Hospital, Paris, France. We report here the clinical course of HAdV-F41 infection and the phylogenetic investigation to identify interpatient transmission. HAdV DNA was quantified in stool and plasma samples by real-time PCR. HAdV type was determined by sequencing of the fiber and hexon genes. Phylogenetic investigation was done with whole-genome sequences obtained by next-generation sequencing. HAdV loads in stool samples ranged from 6.60 to 10.10 log10 copies/ml. HAdV-F41 detection in plasma was observed in four patients, but no disseminated disease was reported. Two patients died, but neither death was attributed to HAdV. While sequencing limited to the fiber gene suggested a cluster with four patients, phylogenetic analysis with whole-genome sequencing (WGS) and HVR7 revealed a cluster that included three patients (C, D, and E), suggesting an interpatient transmission in that cluster and two other independent infections. HAdV-F41 levels in stool specimens of pediatric HSCT patients are high and represent a risk of interpatient transmission. WGS helped to identify related cases. Prompt detection of HAdV in stool and control measures are warranted to limit any risk of nosocomial transmission.
Keywords: WGS; adenovirus; adenovirus disease; hematopoietic stem cell transplantation; next-generation sequencing; outbreak; phylogeny; plasma; stools; whole genome.
Copyright © 2021 American Society for Microbiology.
Figures


Similar articles
-
Adenovirus F41 infection and liver cytolysis in adult hematopoietic stem cell transplant recipients.J Med Virol. 2023 Jul;95(7):e28922. doi: 10.1002/jmv.28922. J Med Virol. 2023. PMID: 37386906
-
A Prolonged Outbreak of Human Adenovirus A31 (HAdV-A31) Infection on a Pediatric Hematopoietic Stem Cell Transplantation Ward with Whole Genome Sequencing Evidence of International Linkages.J Clin Microbiol. 2022 Nov 16;60(11):e0066522. doi: 10.1128/jcm.00665-22. Epub 2022 Oct 12. J Clin Microbiol. 2022. PMID: 36222515 Free PMC article.
-
Adeno-Associated Virus 2 and Human Adenovirus F41 in Wastewater during Outbreak of Severe Acute Hepatitis in Children, Ireland.Emerg Infect Dis. 2023 Apr;29(4):751-760. doi: 10.3201/eid2904.221878. Emerg Infect Dis. 2023. PMID: 36957994 Free PMC article.
-
Pathogenicity and virulence of human adenovirus F41: Possible links to severe hepatitis in children.Virulence. 2023 Dec;14(1):2242544. doi: 10.1080/21505594.2023.2242544. Virulence. 2023. PMID: 37543996 Free PMC article. Review.
-
Current status of human adenovirus infection in China.World J Pediatr. 2022 Aug;18(8):533-537. doi: 10.1007/s12519-022-00568-8. Epub 2022 Jun 18. World J Pediatr. 2022. PMID: 35716276 Free PMC article. Review.
Cited by
-
Multiplex MinION sequencing suggests enteric adenovirus F41 genetic diversity comparable to pre-COVID-19 era.Microb Genom. 2023 Jan;9(1):mgen000920. doi: 10.1099/mgen.0.000920. Microb Genom. 2023. PMID: 36748435 Free PMC article.
-
Isolation of a recombinant simian adenovirus encoding the human adenovirus G52 hexon suggests a simian origin for human adenovirus G52.J Virol. 2024 Apr 16;98(4):e0004324. doi: 10.1128/jvi.00043-24. Epub 2024 Mar 18. J Virol. 2024. PMID: 38497664 Free PMC article.
-
Dissemination and genome characterization of a human adenovirus F41 in a patient with B-Cell lymphoma.Virol J. 2023 Jul 6;20(1):141. doi: 10.1186/s12985-023-02101-3. Virol J. 2023. PMID: 37415207 Free PMC article.
-
Adenovirus 41 diversity in Arizona (USA) using wastewater-based epidemiology, long-range PCR, and pathogen sequencing between October 2019 and March 2020.Epidemiol Infect. 2024 Nov 18;152:e142. doi: 10.1017/S095026882400133X. Epidemiol Infect. 2024. PMID: 39552136 Free PMC article.
-
Molecular phylogeny of human adenovirus type 41 lineages.Virus Evol. 2022 Oct 20;8(2):veac098. doi: 10.1093/ve/veac098. eCollection 2022. Virus Evol. 2022. PMID: 36381230 Free PMC article.
References
-
- Lion T, Kosulin K, Landlinger C, Rauch M, Preuner S, Jugovic D, Pötschger U, Lawitschka A, Peters C, Fritsch G, Matthes-Martin S. 2010. Monitoring of adenovirus load in stool by real-time PCR permits early detection of impending invasive infection in patients after allogeneic stem cell transplantation. Leukemia 24:706–714. 10.1038/leu.2010.4. - DOI - PubMed
-
- Feghoul L, Chevret S, Cuinet A, Dalle J-H, Ouachée M, Yacouben K, Fahd M, Guérin-El Khourouj V, Roupret-Serzec J, Sterkers G, Baruchel A, Simon F, LeGoff J. 2015. Adenovirus infection and disease in paediatric haematopoietic stem cell transplant patients: clues for antiviral pre-emptive treatment. Clin Microbiol Infect 21:701–709. 10.1016/j.cmi.2015.03.011. - DOI - PubMed
-
- Mynarek M, Ganzenmueller T, Mueller-Heine A, Mielke C, Gonnermann A, Beier R, Sauer M, Eiz-Vesper B, Kohstall U, Sykora K-W, Heim A, Maecker-Kolhoff B. 2014. Patient, virus, and treatment-related risk factors in pediatric adenovirus infection after stem cell transplantation: results of a routine monitoring program. Biol Blood Marrow Transplant 20:250–256. 10.1016/j.bbmt.2013.11.009. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

