HIV-Infected Macrophages Are Infected and Killed by the Interferon-Sensitive Rhabdovirus MG1
- PMID: 33568507
- PMCID: PMC8104113
- DOI: 10.1128/JVI.01953-20
HIV-Infected Macrophages Are Infected and Killed by the Interferon-Sensitive Rhabdovirus MG1
Abstract
The use of unique cell surface markers to target and eradicate HIV-infected cells has been a longstanding objective of HIV-1 cure research. This approach, however, overlooks the possibility that intracellular changes present within HIV-infected cells may serve as valuable therapeutic targets. For example, the identification of dysregulated antiviral signaling in cancer has led to the characterization of oncolytic viruses capable of preferentially killing cancer cells. Since impairment of cellular antiviral machinery has been proposed as a mechanism by which HIV-1 evades immune clearance, we hypothesized that HIV-infected macrophages (an important viral reservoir in vivo) would be preferentially killed by the interferon-sensitive oncolytic Maraba virus MG1. We first showed that HIV-infected monocyte-derived macrophages (MDM) were more susceptible to MG1 infection and killing than HIV-uninfected cells. As MG1 is highly sensitive to type I interferons (IFN-I), we then investigated whether we could identify IFN-I signaling differences between HIV-infected and uninfected MDM and found evidence of impaired IFN-α responsiveness within HIV-infected cells. Finally, to assess whether MG1 could target a relevant, primary cell reservoir of HIV-1, we investigated its effects in alveolar macrophages (AM) obtained from effectively treated individuals living with HIV-1. As observed with in vitro-infected MDM, we found that HIV-infected AM were preferentially eliminated by MG1. In summary, the oncolytic rhabdovirus MG1 appears to preferentially target and kill HIV-infected cells via impairment of antiviral signaling pathways and may therefore provide a novel approach to an HIV-1 cure.IMPORTANCE Human immunodeficiency virus type 1 (HIV-1) remains a treatable, but incurable, viral infection. The establishment of viral reservoirs containing latently infected cells remains the main obstacle in the search for a cure. Cure research has also focused on only one cellular target of HIV-1 (the CD4+ T cell) while largely overlooking others (such as macrophages) that contribute to HIV-1 persistence. In this study, we address these challenges by describing a potential strategy for the eradication of HIV-infected macrophages. Specifically, we show that an engineered rhabdovirus-initially developed as a cancer therapy-is capable of preferential infection and killing of HIV-infected macrophages, possibly via the same altered antiviral signaling seen in cancer cells. As this rhabdovirus is currently being explored in phase I/II clinical trials, there is potential for this approach to be readily adapted for use within the HIV-1 cure field.
Keywords: HIV; MG1; interferon; macrophage; oncolytic virus.
Copyright © 2021 Sandstrom et al.
Figures
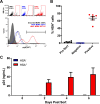
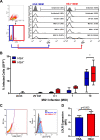

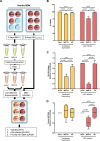
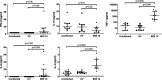
Similar articles
-
The Oncolytic Virus MG1 Targets and Eliminates Cells Latently Infected With HIV-1: Implications for an HIV Cure.J Infect Dis. 2018 Feb 14;217(5):721-730. doi: 10.1093/infdis/jix639. J Infect Dis. 2018. PMID: 29228368 Free PMC article.
-
Brain Macrophages in Simian Immunodeficiency Virus-Infected, Antiretroviral-Suppressed Macaques: a Functional Latent Reservoir.mBio. 2017 Aug 15;8(4):e01186-17. doi: 10.1128/mBio.01186-17. mBio. 2017. PMID: 28811349 Free PMC article.
-
Identification of genetically modified Maraba virus as an oncolytic rhabdovirus.Mol Ther. 2010 Aug;18(8):1440-9. doi: 10.1038/mt.2010.103. Epub 2010 Jun 15. Mol Ther. 2010. PMID: 20551913 Free PMC article.
-
Antiretroviral therapy in macrophages: implication for HIV eradication.Antivir Chem Chemother. 2009 Oct 19;20(2):63-78. doi: 10.3851/IMP1374. Antivir Chem Chemother. 2009. PMID: 19843977 Free PMC article. Review.
-
HIV Persistence in Adipose Tissue Reservoirs.Curr HIV/AIDS Rep. 2018 Feb;15(1):60-71. doi: 10.1007/s11904-018-0378-z. Curr HIV/AIDS Rep. 2018. PMID: 29423731 Free PMC article. Review.
Cited by
-
SMAC Mimetics as Therapeutic Agents in HIV Infection.Front Immunol. 2021 Nov 26;12:780400. doi: 10.3389/fimmu.2021.780400. eCollection 2021. Front Immunol. 2021. PMID: 34899741 Free PMC article. Review.
References
-
- Micci L, Alvarez X, Iriele RI, Ortiz AM, Ryan ES, McGary CS, Deleage C, McAtee BB, He T, Apetrei C, Easley K, Pahwa S, Collman RG, Derdeyn CA, Davenport MP, Estes JD, Silvestri G, Lackner AA, Paiardini M. 2014. CD4 depletion in SIV-infected macaques results in macrophage and microglia infection with rapid turnover of infected cells. PLoS Pathog 10:e1004467. 10.1371/journal.ppat.1004467. - DOI - PMC - PubMed
-
- Abreu CM, Veenhuis RT, Avalos CR, Graham S, Queen SE, Shirk EN, Bullock BT, Li M, Metcalf Pate KA, Beck SE, Mangus LM, Mankowski JL, Clements JE, Gama L. 2019. Infectious virus persists in CD4+ T cells and macrophages in antiretroviral therapy-suppressed simian immunodeficiency virus-infected macaques. J Virol 93:e00065-19. 10.1128/JVI.00065-19. - DOI - PMC - PubMed
-
- Abreu CM, Veenhuis RT, Avalos CR, Graham S, Parrilla DR, Ferreira EA, Queen SE, Shirk EN, Bullock BT, Li M, Metcalf Pate KA, Beck SE, Mangus LM, Mankowski JL, Mac Gabhann F, O’Connor SL, Gama L, Clements JE. 2019. Myeloid and CD4 T cells comprise the latent reservoir in antiretroviral therapy-suppressed SIVmac251-infected macaques. mBio 10:e01659-19. 10.1128/mBio.01659-19. - DOI - PMC - PubMed
-
- Ganor Y, Real F, Sennepin A, Dutertre C-A, Prevedel L, Xu L, Tudor D, Charmeteau B, Couëdel-Courteille A, Marion S, Zenak A-R, Jourdain J-P, Zhou Z, Schmitt A, Capron C, Eugenin EA, Cheynier R, Revol M, Cristofari S, Hosmalin A, Bomsel M. 2019. HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat Microbiol 4:633–644. 10.1038/s41564-018-0335-z. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

