Solid tumours hijack the histone variant network
- PMID: 33568791
- PMCID: PMC8092022
- DOI: 10.1038/s41568-020-00330-0
Solid tumours hijack the histone variant network
Abstract
Cancer is a complex disease characterized by loss of cellular homeostasis through genetic and epigenetic alterations. Emerging evidence highlights a role for histone variants and their dedicated chaperones in cancer initiation and progression. Histone variants are involved in processes as diverse as maintenance of genome integrity, nuclear architecture and cell identity. On a molecular level, histone variants add a layer of complexity to the dynamic regulation of transcription, DNA replication and repair, and mitotic chromosome segregation. Because these functions are critical to ensure normal proliferation and maintenance of cellular fate, cancer cells are defined by their capacity to subvert them. Hijacking histone variants and their chaperones is emerging as a common means to disrupt homeostasis across a wide range of cancers, particularly solid tumours. Here we discuss histone variants and histone chaperones as tumour-promoting or tumour-suppressive players in the pathogenesis of cancer.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
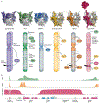

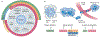
References
-
- Buschbeck M & Hake SB Variants of core histones and their roles in cell fate decisions, development and cancer. Nat. Rev. Mol. Cell Bio 18, 299–314(2017). - PubMed
-
- Talbert PB & Henikoff S Histone variants on the move: substrates for chromatin dynamics. Nat. Rev. Mol. Cell Bio 18, 115–126 (2017). - PubMed
-
- Filipescu D, Müller S & Almouzni G Histone H3 variants and their chaperones during development and disease: contributing to epigenetic control. Annu. Rev. Cell Dev. Biol. 30, 615–646 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

