Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program
- PMID: 33568819
- PMCID: PMC7875770
- DOI: 10.1038/s41586-021-03205-y
Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program
Abstract
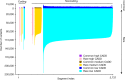
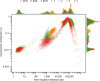
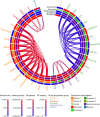
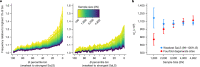
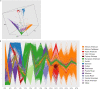
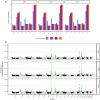
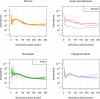
The Trans-Omics for Precision Medicine (TOPMed) programme seeks to elucidate the genetic architecture and biology of heart, lung, blood and sleep disorders, with the ultimate goal of improving diagnosis, treatment and prevention of these diseases. The initial phases of the programme focused on whole-genome sequencing of individuals with rich phenotypic data and diverse backgrounds. Here we describe the TOPMed goals and design as well as the available resources and early insights obtained from the sequence data. The resources include a variant browser, a genotype imputation server, and genomic and phenotypic data that are available through dbGaP (Database of Genotypes and Phenotypes)1. In the first 53,831 TOPMed samples, we detected more than 400 million single-nucleotide and insertion or deletion variants after alignment with the reference genome. Additional previously undescribed variants were detected through assembly of unmapped reads and customized analysis in highly variable loci. Among the more than 400 million detected variants, 97% have frequencies of less than 1% and 46% are singletons that are present in only one individual (53% among unrelated individuals). These rare variants provide insights into mutational processes and recent human evolutionary history. The extensive catalogue of genetic variation in TOPMed studies provides unique opportunities for exploring the contributions of rare and noncoding sequence variants to phenotypic variation. Furthermore, combining TOPMed haplotypes with modern imputation methods improves the power and reach of genome-wide association studies to include variants down to a frequency of approximately 0.01%.
Conflict of interest statement
S.D. holds equity in 23andMe. S.A. holds equity in 23andMe. R.G.B. has received funding from NIH, the COPD Foundation and Alpha1 Foundation. J.F.C. is an inventor on a patent licensed to ImmunArray. M.H.C. has received grant support from GSK. D.L.D. has received personal fees from Novartis. P.T.E. is supported by a grant from Bayer to the Broad Institute focused on the genetics and therapeutics of cardiovascular diseases. P.T.E. has also served on advisory boards or consulted for Quest Diagnostics and Novartis. M.T.G. is a co-inventor on pending patent applications and planned patents directed to the use of recombinant neuroglobin and haeme-based molecules as antidotes for CO poisoning, which have been licensed by Globin Solutions. Globin Solutions also has an option to a potential therapeutic for CO poisoning from VCU, hydroxycobalamin. M.T.G. is a shareholder, advisor and director in Globin Solutions. M.T.G. is a co-inventor on patents directed to the use of nitrite salts in cardiovascular diseases, which were previously licensed to United Therapeutics and Hope Pharmaceuticals, and are now licensed to Globin Solutions. M.T.G. is a co-investigator in a research collaboration with Bayer Pharmaceuticals to evaluate riociguate as a treatment for patients with sickle cell disease. M.T.G. has served as a consultant for Epizyme, Actelion Clinical Research, Acceleron Pharma, Catalyst Biosciences, Modus Therapeutics, Sujana Biotech and United Therapeutics Corporation. M.T.G. is on Bayer HealthCare’s Heart and Vascular Disease Research Advisory Board. D.P.K. receives grants to his institution from Amgen and Radius Health, and serves on scientific advisory boards for Solarea Bio and Pfizer. K.H.L. holds equity in 23andMe. S.A.L. receives sponsored research support from Bristol Myers Squibb/Pfizer, Bayer, Boehringer Ingelheim and Fitbit, has consulted for Bristol Myers Squibb/Pfizer and Bayer, and participates in a research collaboration with IBM. D.D.M. receives research support from Bristol Myers Squibb, Care Evolution, Samsung, Apple Computer, Pfizer, Biotronik, Boehringer Ingelheim, Philips Research Institute, Flexcon, Fitbit and has consulted for Bristol Myers Squibb, Pfizer, Fitbit, Philips, Samsung Electronics, Rose Consulting, Boston Biomedical Associates and FlexCon. D.D.M. is also a member of the Operations Committee and Steering Committee for the GUARD-AF Study (NCT04126486) sponsored by Bristol Meyers Squibb and Pfizer. J.B.M. is an Academic Associate for Quest Diagnostics. For B.D.M.: the Amish Research Program receives partial support from Regeneron Pharmaceuticals. M.E.M. is an inventor on a patent that was published by the United States Patent and Trademark Office on 6 December 2018 under Publication Number US 2018-0346888, and an international patent application that was published on 13 December 2018 under Publication Number WO-2018/226560 regarding B4GALT1 Variants And Uses Thereof. P.N. reports grants from Amgen, Apple, Boston Scientific and Novartis, consulting income from Apple, Blackstone Life Sciences, Genentech and Novartis, and spousal employment at Vertex, all unrelated to the present work. B.M.P. serves on the DSMB of a clinical trial funded by the manufacturer (Zoll LifeCor) and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. J.-S.S. serves as the chairman of Macrogen. S.T.W. is paid royalties by UpToDate. The spouse of C.J.W. works at Regeneron Pharmaceuticals. R.A.G. is an employee of Baylor College of Medicine that receives revenue from Genetic Testing. E.K.S. in the past three years received grant support from GlaxoSmithKline and Bayer. M.C.Z. owns stock in ThermoFisher and Merck. L.A.C. spends part of her time consulting for Dyslipidemia Foundation, a non-profit company, as a statistical consultant. G.R.A. is an employee of Regeneron Pharmaceuticals, he owns stock and stock options for Regeneron Pharmaceuticals.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- R35 HG010692/HG/NHGRI NIH HHS/United States
- P30 ES010126/ES/NIEHS NIH HHS/United States
- U19 CA203654/CA/NCI NIH HHS/United States
- P50 HL118006/HL/NHLBI NIH HHS/United States
- T32 HG000040/HG/NHGRI NIH HHS/United States
- R01 HL090620/HL/NHLBI NIH HHS/United States
- K08 HL141601/HL/NHLBI NIH HHS/United States
- K24 HL148521/HL/NHLBI NIH HHS/United States
- R01 HL111314/HL/NHLBI NIH HHS/United States
- R01 AI132476/AI/NIAID NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- R01 HL131565/HL/NHLBI NIH HHS/United States
- P01 HL132825/HL/NHLBI NIH HHS/United States
- R01 DA044283/DA/NIDA NIH HHS/United States
- R01 HL123915/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- R03 HL141439/HL/NHLBI NIH HHS/United States
- K01 AG059898/AG/NIA NIH HHS/United States
- R01 HL155742/HL/NHLBI NIH HHS/United States
- U01 HL120393/HL/NHLBI NIH HHS/United States
- R01 DK117445/DK/NIDDK NIH HHS/United States
- UH3 HL151865/HL/NHLBI NIH HHS/United States
- R01 HL163972/HL/NHLBI NIH HHS/United States
- R01 AR072199/AR/NIAMS NIH HHS/United States
- R01 HL142711/HL/NHLBI NIH HHS/United States
- I01 BX005295/BX/BLRD VA/United States
- R01 HL113326/HL/NHLBI NIH HHS/United States
- U01 HL137162/HL/NHLBI NIH HHS/United States
- R01 HG005701/HG/NHGRI NIH HHS/United States
- R01 GM075091/GM/NIGMS NIH HHS/United States
- T32 HL007085/HL/NHLBI NIH HHS/United States
- R01 DA037904/DA/NIDA NIH HHS/United States
- R21 HL123677/HL/NHLBI NIH HHS/United States
- P30 DK020572/DK/NIDDK NIH HHS/United States
- R03 HL154284/HL/NHLBI NIH HHS/United States
- R01 MD012765/MD/NIMHD NIH HHS/United States
- R01 HL151855/HL/NHLBI NIH HHS/United States
- U01 HG009088/HG/NHGRI NIH HHS/United States
- UM1 DK078616/DK/NIDDK NIH HHS/United States
- UG3 HL151865/HL/NHLBI NIH HHS/United States
- U01 CA182913/CA/NCI NIH HHS/United States
- T32 CA154274/CA/NCI NIH HHS/United States
- R01 HL149836/HL/NHLBI NIH HHS/United States
- K01 HL135405/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

