Notch signaling: Its essential roles in bone and craniofacial development
- PMID: 33569510
- PMCID: PMC7859553
- DOI: 10.1016/j.gendis.2020.04.006
Notch signaling: Its essential roles in bone and craniofacial development
Abstract
Notch is a cell-cell signaling pathway that is involved in a host of activities including development, oncogenesis, skeletal homeostasis, and much more. More specifically, recent research has demonstrated the importance of Notch signaling in osteogenic differentiation, bone healing, and in the development of the skeleton. The craniofacial skeleton is complex and understanding its development has remained an important focus in biology. In this review we briefly summarize what recent research has revealed about Notch signaling and the current understanding of how the skeleton, skull, and face develop. We then discuss the crucial role that Notch plays in both craniofacial development and the skeletal system, and what importance it may play in the future.
Keywords: Alagille syndrome; Bone; Craniofacial development; Craniosynostosis; Notch; Oncogenesis; Osteogenesis; Spondylocostal dysosotosis.
© 2020 Chongqing Medical University. Production and hosting by Elsevier B.V.
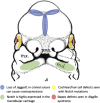
Figures



References
-
- Morgan T.H. The theory of the gene. Am Nat. 1917;51(609):513–544.
-
- Bray S.J. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7(9):678–689. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

