Circular RNA profiles and the potential involvement of down-expression of hsa_circ_0001360 in cutaneous squamous cell carcinogenesis
- PMID: 33569895
- PMCID: PMC8016141
- DOI: 10.1002/2211-5463.13114
Circular RNA profiles and the potential involvement of down-expression of hsa_circ_0001360 in cutaneous squamous cell carcinogenesis
Abstract
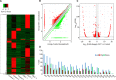
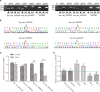
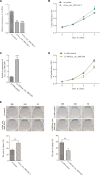
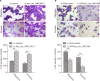
Circular RNAs (circRNAs) act as sponges of noncoding RNAs and have been implicated in many pathophysiological processes, including tumor development and progression. However, their roles in cutaneous squamous cell carcinoma (cSCC) are not yet well understood. This study aimed to identify differentially expressed circRNAs and their potential functions in cutaneous squamous cell carcinogenesis. The expression profiles of circRNAs in three paired cSCC and adjacent nontumorous tissues were detected with RNA sequencing and bioinformatics analysis. The candidate circRNAs were validated by PCR, Sanger sequencing and quantitative RT-PCR in another five matched samples. The biological functions of circRNAs in SCL-1 cells were assessed using circRNA silencing and overexpression, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt (MTS), flow cytometry, transwell and colony formation assays. In addition, the circRNA-miRNA-mRNA interaction networks were predicted by bioinformatics. In summary, 1115 circRNAs, including 457 up-regulated and 658 down-regulated circRNAs (fold change ≥ 2 and P < 0.05), were differentially expressed in cSCC compared with adjacent nontumorous tissues. Of four selected circRNAs, two circRNAs (hsa_circ_0000932 and hsa_circ_0001360) were confirmed to be significantly decreased in cSCC using PCR, Sanger sequencing and quantitative RT-PCR. Furthermore, hsa_circ_0001360 silencing was found to result in a significant increase of the proliferation, migration and invasion but a significant decrease of apoptosis in SCL-1 cells in vitro, whereas hsa_circ_0001360 overexpression showed the opposite regulatory effects. hsa_circ_0001360 was predicted to interact with five miRNAs and their corresponding genes. In conclusion, circRNA dysregulation may play a critical role in carcinogenesis of cSCC, and hsa_circ_0001360 may have potential as a biomarker for cSCC.
Keywords: circular RNA; cutaneous squamous cell carcinoma; expression profile; hsa_circ_0001360.
© 2021 The Authors. FEBS Open Bio published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Que SKT, Zwald FO and Schmults CD (2018) Cutaneous squamous cell carcinoma: incidence, risk factors, diagnosis, and staging. J Am Acad Dermatol 78, 237–247. - PubMed
-
- Karia PS, Han J and Schmults CD (2013) Cutaneous squamous cell carcinoma: estimated incidence of disease, nodal metastasis, and deaths from disease in the United States, 2012. J Am Acad Dermatol 68, 957–966. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

