Computational analyses reveal spatial relationships between nuclear pore complexes and specific lamins
- PMID: 33570570
- PMCID: PMC7883741
- DOI: 10.1083/jcb.202007082
Computational analyses reveal spatial relationships between nuclear pore complexes and specific lamins
Abstract
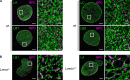
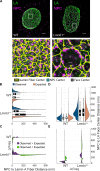
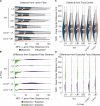
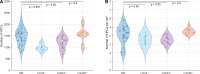
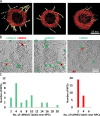
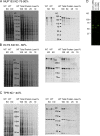
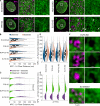
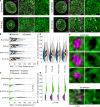
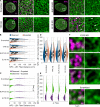
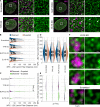
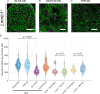
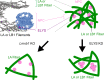
Nuclear lamin isoforms form fibrous meshworks associated with nuclear pore complexes (NPCs). Using datasets prepared from subpixel and segmentation analyses of 3D-structured illumination microscopy images of WT and lamin isoform knockout mouse embryo fibroblasts, we determined with high precision the spatial association of NPCs with specific lamin isoform fibers. These relationships are retained in the enlarged lamin meshworks of Lmna-/- and Lmnb1-/- fibroblast nuclei. Cryo-ET observations reveal that the lamin filaments composing the fibers contact the nucleoplasmic ring of NPCs. Knockdown of the ring-associated nucleoporin ELYS induces NPC clusters that exclude lamin A/C fibers but include LB1 and LB2 fibers. Knockdown of the nucleoporin TPR or NUP153 alters the arrangement of lamin fibers and NPCs. Evidence that the number of NPCs is regulated by specific lamin isoforms is presented. Overall the results demonstrate that lamin isoforms and nucleoporins act together to maintain the normal organization of lamin meshworks and NPCs within the nuclear envelope.
© 2021 Kittisopikul et al.
Figures







References
-
- Broers, J.L., Machiels B.M., van Eys G.J., Kuijpers H.J., Manders E.M., van Driel R., and Ramaekers F.C.. 1999. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J. Cell Sci. 112:3463–3475 https://jcs.biologists.org/content/112/20/3463. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

