Comparison of Severe Acute Respiratory Syndrome Coronavirus 2 Screening Using Reverse Transcriptase-Quantitative Polymerase Chain Reaction or CRISPR-Based Assays in Asymptomatic College Students
- PMID: 33570576
- PMCID: PMC7879237
- DOI: 10.1001/jamanetworkopen.2020.37129
Comparison of Severe Acute Respiratory Syndrome Coronavirus 2 Screening Using Reverse Transcriptase-Quantitative Polymerase Chain Reaction or CRISPR-Based Assays in Asymptomatic College Students
Abstract
Importance: The reopening of colleges and universities in the US during the coronavirus disease 2019 (COVID-19) pandemic is a significant public health challenge. The development of accessible and practical approaches for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection in the college population is paramount for deploying recurrent surveillance testing as an essential strategy for virus detection, containment, and mitigation.
Objective: To determine the prevalence of SARS-CoV-2 in asymptomatic participants in a university community by using CREST (Cas13-based, rugged, equitable, scalable testing), a CRISPR-based test developed for accessible and large-scale viral screening.
Design, setting, and participants: For this cohort study, a total of 1808 asymptomatic participants were screened for SARS-CoV-2 using a CRISPR-based assay and a point-of-reference reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) test. Viral prevalence in self-collected oropharyngeal swab samples collected from May 28 to June 11, 2020, and from June 23 to July 2, 2020, was evaluated.
Exposures: Testing for SARS-CoV-2.
Main outcomes and measures: SARS-CoV-2 status, viral load, and demographic information of the study participants were collected.
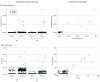
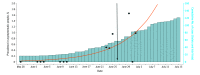
Results: Among the 1808 participants (mean [SD] age, 27.3 [11.0] years; 955 [52.8%] female), 732 underwent testing from May to early June (mean [SD] age, 28.4 [11.7] years; 392 [53.6%] female). All test results in this cohort were negative. In contrast, 1076 participants underwent testing from late June to early July (mean [SD] age, 26.6 [10.5] years; 563 [52.3%] female), with 9 positive results by RT-qPCR. Eight of these positive samples were detected by the CRISPR-based assay and confirmed by Clinical Laboratory Improvement Amendments-certified diagnostic testing. The mean (SD) age of the positive cases was 21.7 (3.3) years; all 8 individuals self-identified as students. These metrics showed that a CRISPR-based assay was effective at capturing positive SARS-CoV-2 cases in this student population. Notably, the viral loads detected in these asymptomatic cases resemble those seen in clinical samples, highlighting the potential of covert viral transmission. The shift in viral prevalence coincided with the relaxation of stay-at-home measures.
Conclusions and relevance: These findings reveal a shift in SARS-CoV-2 prevalence in a young and asymptomatic population and uncover the leading edge of a local outbreak that coincided with rising case counts in the surrounding county and the state of California. The concordance between CRISPR-based and RT-qPCR testing suggests that CRISPR-based assays are reliable and offer alternative options for surveillance testing and detection of SARS-CoV-2 outbreaks, as is required to resume operations in higher-education institutions in the US and abroad.
Conflict of interest statement
Figures

References
-
- Ferguson NM, Laydon D, Nedjati-Gilani G, et al. Impact of non-pharmaceutical interventions (NPIs) to reduce COVID-19 mortality and healthcare demand. Imperial College London. Published March 16, 2020. Accessed January 8, 2021. https://www.imperial.ac.uk/media/imperial-college/medicine/sph/ide/gida-...
-
- Centers for Disease Control and Prevention United States COVID-19 cases and deaths by state. Published 2020. Accessed September 30, 2020. https://www.cdc.gov/covid-data-tracker/
-
- National Center for Education Statistics The Condition of Education: Undergraduate Enrollment. Postsecondary Students Institute of Education Sciences; 2020. Accessed September 30, 2020. https://nces.ed.gov/programs/coe/indicator_cha.asp
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

