Validation of a deep-learning semantic segmentation approach to fully automate MRI-based left-ventricular deformation analysis in cardiotoxicity
- PMID: 33571002
- PMCID: PMC8010548
- DOI: 10.1259/bjr.20201101
Validation of a deep-learning semantic segmentation approach to fully automate MRI-based left-ventricular deformation analysis in cardiotoxicity
Abstract
Objective: Left-ventricular (LV) strain measurements with the Displacement Encoding with Stimulated Echoes (DENSE) MRI sequence provide accurate estimates of cardiotoxicity damage related to chemotherapy for breast cancer. This study investigated an automated and supervised deep convolutional neural network (DCNN) model for LV chamber quantification before strain analysis in DENSE images.
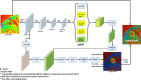
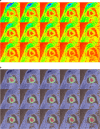
Methods: The DeepLabV3 +DCNN with three versions of ResNet-50 backbone was designed to conduct chamber quantification on 42 female breast cancer data sets. The convolutional layers in the three ResNet-50 backbones were varied as non-atrous, atrous and modified, atrous with accuracy improvements like using Laplacian of Gaussian filters. Parameters such as LV end-diastolic diameter (LVEDD) and ejection fraction (LVEF) were quantified, and myocardial strains analyzed with the Radial Point Interpolation Method (RPIM). Myocardial classification was validated with the performance metrics of accuracy, Dice, average perpendicular distance (APD) and others. Repeated measures ANOVA and intraclass correlation (ICC) with Cronbach's α (C-Alpha) tests were conducted between the three DCNNs and a vendor tool on chamber quantification and myocardial strain analysis.
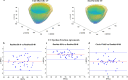
Results: Validation results in the same test-set for myocardial classification were accuracy = 97%, Dice = 0.92, APD = 1.2 mm with the modified ResNet-50, and accuracy = 95%, Dice = 0.90, APD = 1.7 mm with the atrous ResNet-50. The ICC results between the modified ResNet-50, atrous ResNet-50 and vendor-tool were C-Alpha = 0.97 for LVEF (55±7%, 54±7%, 54±7%, p = 0.6), and C-Alpha = 0.87 for LVEDD (4.6 ± 0.3 cm, 4.6 ± 0.3 cm, 4.6 ± 0.4 cm, p = 0.7).
Conclusion: Similar performance metrics and equivalent parameters obtained from comparisons between the atrous networks and vendor tool show that segmentation with the modified, atrous DCNN is applicable for automated LV chamber quantification and subsequent strain analysis in cardiotoxicity.
Advances in knowledge: A novel deep-learning technique for segmenting DENSE images was developed and validated for LV chamber quantification and strain analysis in cardiotoxicity detection.
Figures



Similar articles
-
A deep-learning semantic segmentation approach to fully automated MRI-based left-ventricular deformation analysis in cardiotoxicity.Magn Reson Imaging. 2021 May;78:127-139. doi: 10.1016/j.mri.2021.01.005. Epub 2021 Feb 8. Magn Reson Imaging. 2021. PMID: 33571634 Free PMC article.
-
Automated segmentation of the left-ventricle from MRI with a fully convolutional network to investigate CTRCD in breast cancer patients.J Med Imaging (Bellingham). 2024 Mar;11(2):024003. doi: 10.1117/1.JMI.11.2.024003. Epub 2024 Mar 19. J Med Imaging (Bellingham). 2024. PMID: 38510543 Free PMC article.
-
Can post-chemotherapy cardiotoxicity be detected in long-term survivors of breast cancer via comprehensive 3D left-ventricular contractility (strain) analysis?Magn Reson Imaging. 2019 Oct;62:94-103. doi: 10.1016/j.mri.2019.06.020. Epub 2019 Jun 27. Magn Reson Imaging. 2019. PMID: 31254595 Free PMC article.
-
Fully automated and comprehensive MRI-based left-ventricular contractility analysis in post-chemotherapy breast cancer patients.Br J Radiol. 2020 Jan;93(1105):20190289. doi: 10.1259/bjr.20190289. Epub 2019 Oct 23. Br J Radiol. 2020. PMID: 31617732 Free PMC article.
-
Direct left-ventricular global longitudinal strain (GLS) computation with a fully convolutional network.J Biomech. 2022 Jan;130:110878. doi: 10.1016/j.jbiomech.2021.110878. Epub 2021 Nov 27. J Biomech. 2022. PMID: 34871894 Free PMC article.
Cited by
-
Machine-Learning-Based Diagnostics of Cardiac Sarcoidosis Using Multi-Chamber Wall Motion Analyses.Diagnostics (Basel). 2023 Jul 20;13(14):2426. doi: 10.3390/diagnostics13142426. Diagnostics (Basel). 2023. PMID: 37510168 Free PMC article.
-
A Machine Learning Challenge: Detection of Cardiac Amyloidosis Based on Bi-Atrial and Right Ventricular Strain and Cardiac Function.Diagnostics (Basel). 2022 Nov 4;12(11):2693. doi: 10.3390/diagnostics12112693. Diagnostics (Basel). 2022. PMID: 36359536 Free PMC article.
-
Machine Learning in Cardio-Oncology: New Insights from an Emerging Discipline.Rev Cardiovasc Med. 2023 Oct 19;24(10):296. doi: 10.31083/j.rcm2410296. eCollection 2023 Oct. Rev Cardiovasc Med. 2023. PMID: 39077576 Free PMC article. Review.
References
-
- Bjorck J, Gomes C, Selman B, Weinberger KQ. Understanding batch normalization in 32nd conference on neural information processing systems (NeurIPS 2018). NIPS; 2018. pp. 180–4.
-
- Chen LC, Barron JT, Papandreou G, Murphy K, Yuille AL. Semantic image segmentation with task-specific edge detection using cnns and a discriminatively trained domain transform. CVPR; 2016. pp. 4545–54.
-
- Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL. DeepLab: semantic image segmentation with deep Convolutional nets, Atrous convolution, and fully connected CRFs. CVPR. 40; 2016. pp. 834–48. - PubMed
-
- Chen LC, Yukun Z, Papandreou G, Schroff F, Hartwig A. Encoder-decoder with Atrous separable convolution for semantic image segmentation. CVPR; 2018. pp. 801–18.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

