EphrinB2 clustering by Nipah virus G is required to activate and trap F intermediates at supported lipid bilayer-cell interfaces
- PMID: 33571127
- PMCID: PMC7840137
- DOI: 10.1126/sciadv.abe1235
EphrinB2 clustering by Nipah virus G is required to activate and trap F intermediates at supported lipid bilayer-cell interfaces
Abstract
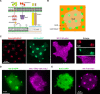
Paramyxovirus membrane fusion requires an attachment protein that binds to a host cell receptor and a fusion protein that merges the viral and host membranes. For Nipah virus (NiV), the G attachment protein binds ephrinB2/B3 receptors and activates F-mediated fusion. To visualize dynamic events of these proteins at the membrane interface, we reconstituted NiV fusion activation by overlaying F- and G-expressing cells onto ephrinB2-functionalized supported lipid bilayers and used TIRF microscopy to follow F, G, and ephrinB2. We found that G and ephrinB2 form clusters and that oligomerization of ephrinB2 is necessary for F activation. Single-molecule tracking of F particles revealed accumulation of an immobilized intermediate upon activation. We found no evidence for stable F-G protein complexes before or after activation. These observations lead to a revised model for NiV fusion activation and provide a foundation for investigating other multicomponent viral fusion systems.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures



References
-
- Kielian M., Mechanisms of virus membrane fusion proteins. Annu. Rev. Virol. 1, 171–189 (2014). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

