Antibody responses to endemic coronaviruses modulate COVID-19 convalescent plasma functionality
- PMID: 33571169
- PMCID: PMC8011893
- DOI: 10.1172/JCI146927
Antibody responses to endemic coronaviruses modulate COVID-19 convalescent plasma functionality
Abstract
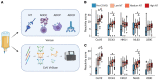
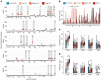
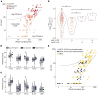
SARS-CoV-2 (CoV2) antibody therapies, including COVID-19 convalescent plasma (CCP), monoclonal antibodies, and hyperimmune globulin, are among the leading treatments for individuals with early COVID-19 infection. The functionality of convalescent plasma varies greatly, but the association of antibody epitope specificities with plasma functionality remains uncharacterized. We assessed antibody functionality and reactivities to peptides across the CoV2 and the 4 endemic human coronavirus (HCoV) genomes in 126 CCP donations. We found strong correlation between plasma functionality and polyclonal antibody targeting of CoV2 spike protein peptides. Antibody reactivity to many HCoV spike peptides also displayed strong correlation with plasma functionality, including pan-coronavirus cross-reactive epitopes located in a conserved region of the fusion peptide. After accounting for antibody cross-reactivity, we identified an association between greater alphacoronavirus NL63 antibody responses and development of highly neutralizing antibodies against CoV2. We also found that plasma preferentially reactive to the CoV2 spike receptor binding domain (RBD), versus the betacoronavirus HKU1 RBD, had higher neutralizing titer. Finally, we developed a 2-peptide serosignature that identifies plasma donations with high anti-spike titer, but that suffer from low neutralizing activity. These results suggest that analysis of coronavirus antibody fine specificities may be useful for selecting desired therapeutics and understanding the complex immune responses elicited by CoV2 infection.
Keywords: Adaptive immunity; Immunology; Infectious disease.
Conflict of interest statement
Figures


Update of
-
Antibody responses to endemic coronaviruses modulate COVID-19 convalescent plasma functionality.medRxiv [Preprint]. 2020 Dec 18:2020.12.16.20248294. doi: 10.1101/2020.12.16.20248294. medRxiv. 2020. Update in: J Clin Invest. 2021 Apr 1;131(7):146927. doi: 10.1172/JCI146927. PMID: 33354688 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI152078/AI/NIAID NIH HHS/United States
- T32 GM136577/GM/NIGMS NIH HHS/United States
- R01 AI120938/AI/NIAID NIH HHS/United States
- U24 AI118633/AI/NIAID NIH HHS/United States
- R01 AI052733/AI/NIAID NIH HHS/United States
- R01 AI128779/AI/NIAID NIH HHS/United States
- R01 NR005228/NR/NINR NIH HHS/United States
- UM1 AI068613/AI/NIAID NIH HHS/United States
- HHSN272201400007C/AI/NIAID NIH HHS/United States
- R01 GM136724/GM/NIGMS NIH HHS/United States
- R01 HL059842/HL/NHLBI NIH HHS/United States
- K23 HL151826/HL/NHLBI NIH HHS/United States
- T32 AI007363/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

