Potential Dual Role of West Nile Virus NS2B in Orchestrating NS3 Enzymatic Activity in Viral Replication
- PMID: 33572517
- PMCID: PMC7911885
- DOI: 10.3390/v13020216
Potential Dual Role of West Nile Virus NS2B in Orchestrating NS3 Enzymatic Activity in Viral Replication
Abstract
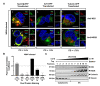
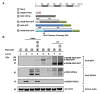
West Nile virus (WNV) nonstructural protein 3 (NS3) harbors the viral triphosphatase and helicase for viral RNA synthesis and, together with NS2B, constitutes the protease responsible for polyprotein processing. NS3 is a soluble protein, but it is localized to specialized compartments at the rough endoplasmic reticulum (RER), where its enzymatic functions are essential for virus replication. However, the mechanistic details behind the recruitment of NS3 from the cytoplasm to the RER have not yet been fully elucidated. In this study, we employed immunofluorescence and biochemical assays to demonstrate that NS3, when expressed individually and when cleaved from the viral polyprotein, is localized exclusively to the cytoplasm. Furthermore, NS3 appeared to be peripherally recruited to the RER and proteolytically active when NS2B was provided in trans. Thus, we provide evidence for a potential additional role for NS2B in not only serving as the cofactor for the NS3 protease, but also in recruiting NS3 from the cytoplasm to the RER for proper enzymatic activity. Results from our study suggest that targeting the interaction between NS2B and NS3 in disrupting the NS3 ER localization may be an attractive avenue for antiviral drug discovery.
Keywords: NS2B; NS3; West Nile Virus; endoplasmic reticulum; flavivirus; membrane structures.
Conflict of interest statement
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Heinz F.X., Collett M.S., Purcell R.H., Gould E.A., Howard C.R., Houghton M., Moormann R.J., Rice C.M., Tiehl H.J. Flaviviridae. In: van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., Carstens E.B., Estes M.K., Lemon S.M., Maniloff J., Mayo M.A., McGeoch D.J., Pringle C.R., et al., editors. Virus taxonomy: Classification and nomenclature of viruses. 7th Report of the International Committee for the Taxonomy of Viruses. Academic Press; San Diego, CA, USA: 2000. pp. 869–878.
-
- Nowak T., Farber P.M., Wengler G., Wengler G. Analyses of the terminal sequences of West Nile virus structural proteins and of the in vitro translation of these proteins allow the proposal of a complete scheme of the proteolytic cleavages involved in their synthesis. Virology. 1989;169:365–376. doi: 10.1016/0042-6822(89)90162-1. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

