Pedigree and Molecular Analyses in the Assessment of Genetic Variability of the Polish Greyhound
- PMID: 33572519
- PMCID: PMC7911804
- DOI: 10.3390/ani11020353
Pedigree and Molecular Analyses in the Assessment of Genetic Variability of the Polish Greyhound
Abstract
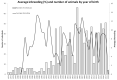
The aim of the study was to assess the genetic variability of the Polish Greyhound population based on pedigree analysis and molecular DNA testing and to determine the degree of relatedness among individuals in the population. Pedigree data of 912 Polish Greyhounds recorded in pedigree books since they were opened for this breed were analyzed. For molecular testing, DNA was obtained from cheek swabs taken from 235 dogs of the tested breed. A panel of 21 markers (Short Tandem Repeat-STR) was used. The mean inbreeding determined for the Polish Greyhound population based on pedigree analyses was low and amounted to 11.8%, but as many as 872 individuals of the 912 dogs in the studied population were inbred. A total of 83 founders (at least one unknown parent) were identified, among which 27 founders had both unknown parents. Full-sibling groups consisted of 130 individuals, with a minimum and maximum litter size of 2 and 16, respectively. The average litter size was 5.969. Gene diversity calculated based on the mean kinship matrix was 0.862 and the population mean kinship was 0.138. The founder genome equivalent based on the mean kinship matrix was 3.61; the founder genome surviving level was 12.34; the mean Ne was estimated at 21.76; and the Ne/N ratio was 0.135. The FIS inbreeding coefficient for 21 STR was negative, and the mean FIS value for all loci had a low negative value (-0.018). These values suggest a low level of inbreeding in the examined breed as well as the avoidance of mating related animals.
Keywords: Polish Greyhound; founders; molecular analysis; pedigree analysis.
Conflict of interest statement
The authors declare to have no conflicts of interest.
Figures



Similar articles
-
Population structure and genetic diversity of Polish Arabian horses based on pedigree data.Animal. 2024 May;18(5):101148. doi: 10.1016/j.animal.2024.101148. Epub 2024 Mar 27. Animal. 2024. PMID: 38642411
-
Genetic Variability and Population Structure of Polish Konik Horse Maternal Lines Based on Microsatellite Markers.Genes (Basel). 2021 Apr 9;12(4):546. doi: 10.3390/genes12040546. Genes (Basel). 2021. PMID: 33918718 Free PMC article.
-
Population structure and genetic variability in the Murrah dairy breed of water buffalo in Brazil accessed via pedigree analysis.Trop Anim Health Prod. 2012 Dec;44(8):1891-7. doi: 10.1007/s11250-012-0153-x. Epub 2012 Apr 26. Trop Anim Health Prod. 2012. PMID: 22535150
-
Pedigree analysis of the Turkish Arab horse population: structure, inbreeding and genetic variability.Animal. 2017 Sep;11(9):1449-1456. doi: 10.1017/S175173111700009X. Epub 2017 Feb 13. Animal. 2017. PMID: 28190413
-
Monitoring of genetic diversity in the endangered Martina Franca donkey population.J Anim Sci. 2011 May;89(5):1304-11. doi: 10.2527/jas.2010-3379. J Anim Sci. 2011. PMID: 21521813
Cited by
-
Genetic Diversity and Trends of Ancestral and New Inbreeding in Deutsch Drahthaar Assessed by Pedigree Data.Animals (Basel). 2022 Apr 5;12(7):929. doi: 10.3390/ani12070929. Animals (Basel). 2022. PMID: 35405917 Free PMC article.
-
Kazakh national dog breed Tazy: What do we know?PLoS One. 2023 Mar 8;18(3):e0282041. doi: 10.1371/journal.pone.0282041. eCollection 2023. PLoS One. 2023. PMID: 36888576 Free PMC article.
-
Microsatellite DNA Analysis of Genetic Diversity and Parentage Testing in the Popular Dog Breeds in Poland.Genes (Basel). 2021 Mar 26;12(4):485. doi: 10.3390/genes12040485. Genes (Basel). 2021. PMID: 33810589 Free PMC article.
References
-
- Szmurło M. Naprawdę Charty Polskie. Pies. 1982;3:7.
-
- FCI Standard and Nomenclature. [(accessed on 23 November 2020)]; Available online: http://www.fci.be/en/nomenclature/POLISH-GREYHOUND-333.html.
-
- Ciampolini R., Cecchi F., Bramante A., Casetti F., Presciuttini S. Genetic variability of the Bracco Italiano dog breed based on microsatellite polymorphism. Ital. J. Anim. Sci. 2011;10:e59. doi: 10.4081/ijas.2011.e59. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

