Identification of an RNA Silencing Suppressor Encoded by a Symptomless Fungal Hypovirus, Cryphonectria Hypovirus 4
- PMID: 33572564
- PMCID: PMC7912522
- DOI: 10.3390/biology10020100
Identification of an RNA Silencing Suppressor Encoded by a Symptomless Fungal Hypovirus, Cryphonectria Hypovirus 4
Abstract
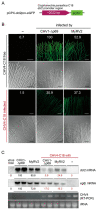
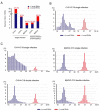
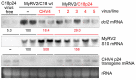
Previously, we have reported the ability of a symptomless hypovirus Cryphonectria hypovirus 4 (CHV4) of the chestnut blight fungus to facilitate stable infection by a co-infecting mycoreovirus 2 (MyRV2)-likely through the inhibitory effect of CHV4 on RNA silencing (Aulia et al., Virology, 2019). In this study, the N-terminal portion of the CHV4 polyprotein, termed p24, is identified as an autocatalytic protease capable of suppressing host antiviral RNA silencing. Using a bacterial expression system, CHV4 p24 is shown to cleave autocatalytically at the di-glycine peptide (Gly214-Gly215) of the polyprotein through its protease activity. Transgenic expression of CHV4 p24 in Cryphonectria parasitica suppresses the induction of one of the key genes of the antiviral RNA silencing, dicer-like 2, and stabilizes the infection of RNA silencing-susceptible virus MyRV2. This study shows functional similarity between CHV4 p24 and its homolog p29, encoded by the symptomatic prototype hypovirus CHV1.
Keywords: Cryphonectria parasitica; Dicer; RNA silencing; RNAi suppressor; chestnut blight fungus; co-infection; hypovirus; mycovirus; reovirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

