Combined Metabolite and Transcriptome Profiling Reveals the Norisoprenoid Responses in Grape Berries to Abscisic Acid and Synthetic Auxin
- PMID: 33572582
- PMCID: PMC7867017
- DOI: 10.3390/ijms22031420
Combined Metabolite and Transcriptome Profiling Reveals the Norisoprenoid Responses in Grape Berries to Abscisic Acid and Synthetic Auxin
Abstract
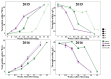
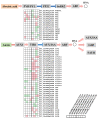
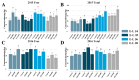
The abscisic acid (ABA) increase and auxin decline are both indicators of ripening initiation in grape berry, and norisoprenoid accumulation also starts at around the onset of ripening. However, the relationship between ABA, auxin, and norisoprenoids remains largely unknown, especially at the transcriptome level. To investigate the transcriptional and posttranscriptional regulation of the ABA and synthetic auxin 1-naphthaleneacetic acid (NAA) on norisoprenoid production, we performed time-series GC-MS and RNA-seq analyses on Vitis vinifera L. cv. Cabernet Sauvignon grape berries from pre-veraison to ripening. Higher levels of free norisoprenoids were found in ABA-treated mature berries in two consecutive seasons, and both free and total norisoprenoids were significantly increased by NAA in one season. The expression pattern of known norisoprenoid-associated genes in all samples and the up-regulation of specific alternative splicing isoforms of VviDXS and VviCRTISO in NAA-treated berries were predicted to contribute to the norisoprenoid accumulation in ABA and NAA-treated berries. Combined weighted gene co-expression network analysis (WGCNA) and DNA affinity purification sequencing (DAP-seq) analysis suggested that VviGATA26, and the previously identified switch genes of myb RADIALIS (VIT_207s0005g02730) and MAD-box (VIT_213s0158g00100) could be potential regulators of norisoprenoid accumulation. The positive effects of ABA on free norisoprenoids and NAA on total norisoprenoid accumulation were revealed in the commercially ripening berries. Since the endogenous ABA and auxin are sensitive to environmental factors, this finding provides new insights to develop viticultural practices for managing norisoprenoids in vineyards in response to changing climates.
Keywords: DAP-seq; abscisic acid; alternative splicing; gene expression; grape berry ripening; naphthaleneacetic acid; norisoprenoid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Timing of ripening initiation in grape berries and its relationship to seed content and pericarp auxin levels.BMC Plant Biol. 2015 Feb 12;15:46. doi: 10.1186/s12870-015-0440-6. BMC Plant Biol. 2015. PMID: 25848949 Free PMC article.
-
Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development.BMC Genomics. 2007 Nov 22;8:429. doi: 10.1186/1471-2164-8-429. BMC Genomics. 2007. PMID: 18034876 Free PMC article.
-
Integrated Analysis of Transcriptome and Metabolome to Unveil Impact on Enhancing Grape Aroma Quality with Synthetic Auxin: Spotlight the Mediation of ABA in Crosstalk with Auxin.J Agric Food Chem. 2024 Jan 17;72(2):1228-1243. doi: 10.1021/acs.jafc.3c06846. Epub 2024 Jan 5. J Agric Food Chem. 2024. PMID: 38181223
-
Complex Interplay of Hormonal Signals during Grape Berry Ripening.Molecules. 2015 May 21;20(5):9326-43. doi: 10.3390/molecules20059326. Molecules. 2015. PMID: 26007186 Free PMC article. Review.
-
Insights into the cell-wall dynamics in grapevine berries during ripening and in response to biotic and abiotic stresses.Plant Mol Biol. 2024 Apr 11;114(3):38. doi: 10.1007/s11103-024-01437-w. Plant Mol Biol. 2024. PMID: 38605193 Free PMC article. Review.
Cited by
-
Effect of Cold Stabilization Duration on Organic Acids and Aroma Compounds during Vitis vinifera L. cv. Riesling Wine Bottle Storage.Foods. 2022 Apr 19;11(9):1179. doi: 10.3390/foods11091179. Foods. 2022. PMID: 35563903 Free PMC article.
-
Composition of Pinot Noir Wine from Grapevine Red Blotch Disease-Infected Vines Managed with Exogenous Abscisic Acid Applications.Molecules. 2022 Jul 15;27(14):4520. doi: 10.3390/molecules27144520. Molecules. 2022. PMID: 35889392 Free PMC article.
-
Identification of SNP loci and candidate genes genetically controlling norisoprenoids in grape berry based on genome-wide association study.Front Plant Sci. 2023 Mar 1;14:1142139. doi: 10.3389/fpls.2023.1142139. eCollection 2023. Front Plant Sci. 2023. PMID: 36938056 Free PMC article.
-
Genetic architecture of berry aroma compounds in a QTL (quantitative trait loci) mapping population of interspecific hybrid grapes (Vitis labruscana × Vitis vinifera).BMC Plant Biol. 2022 Sep 23;22(1):458. doi: 10.1186/s12870-022-03842-z. BMC Plant Biol. 2022. PMID: 36151514 Free PMC article.
-
Secondary metabolites in grapevine: crosstalk of transcriptional, metabolic and hormonal signals controlling stress defence responses in berries and vegetative organs.Front Plant Sci. 2023 Jun 19;14:1124298. doi: 10.3389/fpls.2023.1124298. eCollection 2023. Front Plant Sci. 2023. PMID: 37404528 Free PMC article. Review.
References
-
- Williams P.J., Sefton M.A., Francis I.L. Glycosidic precursors of varietal grape and wine flavor. In: Teranishi R., Takeoka G.R., Günterm M., editors. Flavour Precursors, Thermal and Enzymatic Conversions. American Chemical Society; Washington, DC, USA: 1992. pp. 74–86. (ACS Symposium Series 490). - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

