Structure, Function, and Interactions of the HIV-1 Capsid Protein
- PMID: 33572761
- PMCID: PMC7910843
- DOI: 10.3390/life11020100
Structure, Function, and Interactions of the HIV-1 Capsid Protein
Abstract
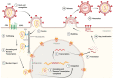
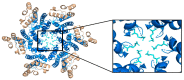
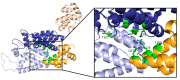
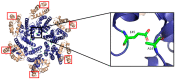
The capsid (CA) protein of the human immunodeficiency virus type 1 (HIV-1) is an essential structural component of a virion and facilitates many crucial life cycle steps through interactions with host cell factors. Capsid shields the reverse transcription complex from restriction factors while it enables trafficking to the nucleus by hijacking various adaptor proteins, such as FEZ1 and BICD2. In addition, the capsid facilitates the import and localization of the viral complex in the nucleus through interaction with NUP153, NUP358, TNPO3, and CPSF-6. In the later stages of the HIV-1 life cycle, CA plays an essential role in the maturation step as a constituent of the Gag polyprotein. In the final phase of maturation, Gag is cleaved, and CA is released, allowing for the assembly of CA into a fullerene cone, known as the capsid core. The fullerene cone consists of ~250 CA hexamers and 12 CA pentamers and encloses the viral genome and other essential viral proteins for the next round of infection. As research continues to elucidate the role of CA in the HIV-1 life cycle and the importance of the capsid protein becomes more apparent, CA displays potential as a therapeutic target for the development of HIV-1 inhibitors.
Keywords: HIV-1/AIDS; assembly; capsid; host proteins; post-entry events; restriction factors; virus-host interactions.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the study’s design, in the analyses or interpretation of data; in the writing of the manuscript, or in the decision to publish the manuscript.
Figures





References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

