The Temporal Order of DNA Replication Shaped by Mammalian DNA Methyltransferases
- PMID: 33572832
- PMCID: PMC7911666
- DOI: 10.3390/cells10020266
The Temporal Order of DNA Replication Shaped by Mammalian DNA Methyltransferases
Abstract
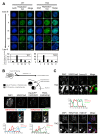
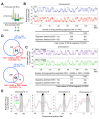
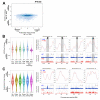
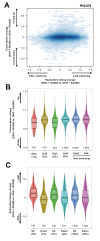
Multiple epigenetic pathways underlie the temporal order of DNA replication (replication timing) in the contexts of development and disease. DNA methylation by DNA methyltransferases (Dnmts) and downstream chromatin reorganization and transcriptional changes are thought to impact DNA replication, yet this remains to be comprehensively tested. Using cell-based and genome-wide approaches to measure replication timing, we identified a number of genomic regions undergoing subtle but reproducible replication timing changes in various Dnmt-mutant mouse embryonic stem (ES) cell lines that included a cell line with a drug-inducible Dnmt3a2 expression system. Replication timing within pericentromeric heterochromatin (PH) was shown to be correlated with redistribution of H3K27me3 induced by DNA hypomethylation: Later replicating PH coincided with H3K27me3-enriched regions. In contrast, this relationship with H3K27me3 was not evident within chromosomal arm regions undergoing either early-to-late (EtoL) or late-to-early (LtoE) switching of replication timing upon loss of the Dnmts. Interestingly, Dnmt-sensitive transcriptional up- and downregulation frequently coincided with earlier and later shifts in replication timing of the chromosomal arm regions, respectively. Our study revealed the previously unrecognized complex and diverse effects of the Dnmts loss on the mammalian DNA replication landscape.
Keywords: DNA methyltransferases; DNA replication; replication timing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

