BBS Proteins Affect Ciliogenesis and Are Essential for Hedgehog Signaling, but Not for Formation of iPSC-Derived RPE-65 Expressing RPE-Like Cells
- PMID: 33572860
- PMCID: PMC7866284
- DOI: 10.3390/ijms22031345
BBS Proteins Affect Ciliogenesis and Are Essential for Hedgehog Signaling, but Not for Formation of iPSC-Derived RPE-65 Expressing RPE-Like Cells
Abstract
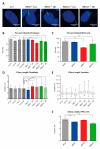
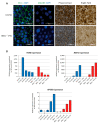
Bardet-Biedl syndrome (BBS) is a ciliopathy characterized by retinal dystrophy, renal cysts, obesity and polydactyly. BBS genes have been implicated in ciliogenesis, hedgehog signaling and retinal pigment epithelium maturation. BBS1 and BBS5 are members of the BBSome, implicated in cilia transport of proteins, and BBS10 is a member of the chaperonin-complex, mediating BBSome assembly. In this study, involvement of BBS1, BBS5 and BBS10 in ciliogenesis and hedgehog signaling were investigated in BBS-defective patient fibroblasts as well as in RPE-hTERT cells following siRNA-mediated knockdown of the BBS genes. Furthermore, the ability of BBS1-defective induced pluripotent stem-cells (iPSCs) to differentiate into RPE cells was assessed. We report that cells lacking functional BBS5 or BBS10 have a reduced number of primary cilia, whereas cells lacking functional BBS1 display shorter primary cilia compared to wild-type cells. Hedgehog signaling was substantially impaired and Smoothened, a component of hedgehog signaling, was trapped inside the cilia of the BBS-defective cells, even in the absence of Smoothened agonist. Preliminary results demonstrated the ability of BBS1-defective iPSC to differentiate into RPE-65 expressing RPE-like cells. The BBS1-/--defective RPE-like cells were less pigmented, compared to RPE-like cells differentiated from control iPSCs, indicating an impact of BBS1 on RPE maturation.
Keywords: BBS; RPE; RPE65; SMO; WNT; ciliogenesis; hedgehog-signaling; induced pluripotent stem cell.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Schaefer E., Stoetzel C., Scheidecker S., Geoffroy V., Prasad M.K., Redin C., Missotte I., Lacombe D., Mandel J.L., Muller J., et al. Identification of a novel mutation confirms the implication of IFT172 (BBS20) in Bardet–Biedl syndrome. J. Hum. Genet. 2016;61:447–450. doi: 10.1038/jhg.2015.162. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

