Imprinted Genes and Multiple Sclerosis: What Do We Know?
- PMID: 33572862
- PMCID: PMC7866243
- DOI: 10.3390/ijms22031346
Imprinted Genes and Multiple Sclerosis: What Do We Know?
Abstract
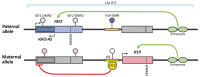
Multiple sclerosis (MS) is a chronic autoimmune neurodegenerative disease of the central nervous system that arises from interplay between non-genetic and genetic risk factors. The epigenetics functions as a link between these factors, affecting gene expression in response to external influence, and therefore should be extensively studied to improve the knowledge of MS molecular mechanisms. Among others, the epigenetic mechanisms underlie the establishment of parent-of-origin effects that appear as phenotypic differences depending on whether the allele was inherited from the mother or father. The most well described manifestation of parent-of-origin effects is genomic imprinting that causes monoallelic gene expression. It becomes more obvious that disturbances in imprinted genes at the least affecting their expression do occur in MS and may be involved in its pathogenesis. In this review we will focus on the potential role of imprinted genes in MS pathogenesis.
Keywords: DLK1-DIO3 locus; genomic imprinting; miRNA; multiple sclerosis; parent-of-origin effect.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Wallin M.T., Culpepper W.J., Nichols E., Bhutta Z.A., Gebrehiwot T.T., Hay S.I., Khalil I.A., Krohn K.J., Liang X., Naghavi M., et al. Global, regional, and national burden of multiple sclerosis 1990–2016: A systematic analysis for the Global Burden of Disease Study. Lancet Neurol. 2019;18:269–285. doi: 10.1016/S1474-4422(18)30443-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

