Integrase Strand Transfer Inhibitors Are Effective Anti-HIV Drugs
- PMID: 33572956
- PMCID: PMC7912079
- DOI: 10.3390/v13020205
Integrase Strand Transfer Inhibitors Are Effective Anti-HIV Drugs
Abstract
Integrase strand transfer inhibitors (INSTIs) are currently recommended for the first line treatment of human immunodeficiency virus type one (HIV-1) infection. The first-generation INSTIs are effective but can select for resistant viruses. Recent advances have led to several potent second-generation INSTIs that are effective against both wild-type (WT) HIV-1 integrase and many of the first-generation INSTI-resistant mutants. The emergence of resistance to these new second-generation INSTIs has been minimal, which has resulted in alternative treatment strategies for HIV-1 patients. Moreover, because of their high antiviral potencies and, in some cases, their bioavailability profiles, INSTIs will probably have prominent roles in pre-exposure prophylaxis (PrEP). Herein, we review the current state of the clinically relevant INSTIs and discuss the future outlook for this class of antiretrovirals.
Keywords: HIV; INSTIs; antiviral therapy; drug resistance; integration.
Conflict of interest statement
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services or the National Institutes of Health, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Figures

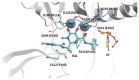
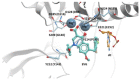
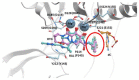
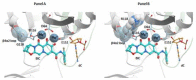
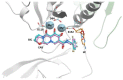

References
-
- U.S. Department of Health and Human Services . Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIV. G-1. The U.S. Department of Health and Human Services Guidelines, AIDSinfo; Washington, DC, USA: 2019.
-
- Cahn P., Madero J.S., Arribas J.R., Antinori A., Ortiz R., Clarke A.E., Hung C.C., Rockstroh J.K., Girard P.M., Sievers J., et al. Dolutegravir plus lamivudine versus dolutegravir plus tenofovir disoproxil fumarate and emtricitabine in antiretroviral-naive adults with HIV-1 infection (GEMINI-1 and GEMINI-2): Week 48 results from two multicentre, double-blind, randomised, non-inferiority, phase 3 trials. Lancet. 2019;393:143–155. doi: 10.1016/S0140-6736(18)32462-0. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

