Moonlighting Proteins Shine New Light on Molecular Signaling Niches
- PMID: 33573037
- PMCID: PMC7866414
- DOI: 10.3390/ijms22031367
Moonlighting Proteins Shine New Light on Molecular Signaling Niches
Abstract
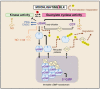
Plants as sessile organisms face daily environmental challenges and have developed highly nuanced signaling systems to enable suitable growth, development, defense, or stalling responses. Moonlighting proteins have multiple tasks and contribute to cellular signaling cascades where they produce additional variables adding to the complexity or fuzziness of biological systems. Here we examine roles of moonlighting kinases that also generate 3',5'-cyclic guanosine monophosphate (cGMP) in plants. These proteins include receptor like kinases and lipid kinases. Their guanylate cyclase activity potentiates the development of localized cGMP-enriched nanodomains or niches surrounding the kinase and its interactome. These nanodomains contribute to allosteric regulation of kinase and other molecules in the immediate complex directly or indirectly modulating signal cascades. Effects include downregulation of kinase activity, modulation of other members of the protein complexes such as cyclic nucleotide gated channels and potential triggering of cGMP-dependent degradation cascades terminating signaling. The additional layers of information provided by the moonlighting kinases are discussed in terms of how they may be used to provide a layer of fuzziness to effectively modulate cellular signaling cascades.
Keywords: 3′,5′-cyclic guanosine monophosphate (cGMP); brassinosteroid insensitive 1 (BRI1); cryptic enzyme; danger associated peptide receptor (PEPR1 and PEPR2); guanylate cyclase; moonlighting proteins; nanodomains; phytosulfokine receptor 1 (PSKR1); receptor like kinase; wall associated kinase like 10 (WAKL10).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Moonlighting kinases with guanylate cyclase activity can tune regulatory signal networks.Plant Signal Behav. 2012 Feb;7(2):201-4. doi: 10.4161/psb.18891. Epub 2012 Feb 1. Plant Signal Behav. 2012. PMID: 22353864 Free PMC article.
-
The brassinosteroid receptor BRI1 can generate cGMP enabling cGMP-dependent downstream signaling.Plant J. 2017 Aug;91(4):590-600. doi: 10.1111/tpj.13589. Epub 2017 Jun 12. Plant J. 2017. PMID: 28482142
-
In Vitro Characterization of Guanylyl Cyclase BdPepR2 from Brachypodium distachyon Identified through a Motif-Based Approach.Int J Mol Sci. 2021 Jun 10;22(12):6243. doi: 10.3390/ijms22126243. Int J Mol Sci. 2021. PMID: 34200573 Free PMC article.
-
Comparison of moonlighting guanylate cyclases: roles in signal direction?Biochem Soc Trans. 2014 Dec;42(6):1773-9. doi: 10.1042/BST20140223. Biochem Soc Trans. 2014. PMID: 25399605 Review.
-
Signal transduction: activation of the guanylate cyclase-cyclic guanosine-3'-5' monophosphate system by hormones and free radicals.Am J Med Sci. 1997 Nov;314(5):311-23. doi: 10.1097/00000441-199711000-00008. Am J Med Sci. 1997. PMID: 9365333 Review.
Cited by
-
Guanylate cyclase activity in moss: revisiting the role of ERECTA-like receptors.Physiol Mol Biol Plants. 2025 May;31(5):813-822. doi: 10.1007/s12298-025-01606-1. Epub 2025 Jun 4. Physiol Mol Biol Plants. 2025. PMID: 40568481 Free PMC article.
-
Modulation of Inflammatory Cytokine Production in Human Monocytes by cGMP and IRAK3.Int J Mol Sci. 2022 Feb 25;23(5):2552. doi: 10.3390/ijms23052552. Int J Mol Sci. 2022. PMID: 35269704 Free PMC article.
-
Intrinsic Disorder and Other Malleable Arsenals of Evolved Protein Multifunctionality.J Mol Evol. 2024 Dec;92(6):669-684. doi: 10.1007/s00239-024-10196-7. Epub 2024 Aug 30. J Mol Evol. 2024. PMID: 39214891 Review.
-
Ca2+ signals in plant immunity.EMBO J. 2022 Jun 14;41(12):e110741. doi: 10.15252/embj.2022110741. Epub 2022 May 13. EMBO J. 2022. PMID: 35560235 Free PMC article. Review.
-
Functional Crypto-Adenylate Cyclases Operate in Complex Plant Proteins.Front Plant Sci. 2021 Aug 12;12:711749. doi: 10.3389/fpls.2021.711749. eCollection 2021. Front Plant Sci. 2021. PMID: 34456950 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

